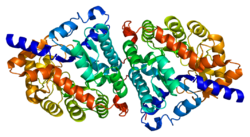
Dipeptidase 1 (DPEP1), or renal dipeptidase, is a membrane-bound glycoprotein responsible for hydrolyzing dipeptides. It is found in the microsomal fraction of the porcine kidney cortex.[5] It exists as a disulfide-linked homodimer that is glygosylphosphatidylinositol (GPI)-anchored to the renal brush border of the kidney.[6] The active site on each homodimer is made up of a barrel subunit with binuclear zinc ions that are bridged by the Gly125 side-chain located at the bottom of the barrel.[7]
Structure
editThe gene coding for DPEP1 is 6 kb long and consists of ten exons and nine introns. The protein itself is made of 411 amino acid residues and is only transcribed in kidney cells.[8] Although disulfide linkages in DPEP1 do not contribute to the enzyme’s activity, they are essential for the enzyme’s proper function because they keep the enzyme’s subunits together and attached to the renal brush border. Cysteine 261 is involved in disulfide linkage between the enzyme’s subunits, and is also located very close to both the site of the GPI-anchor and the membrane, suggesting that it is also involved in the enzyme’s linkage to the membrane.[9]
DPEP1 is also a metalloenzyme that specifically uses zinc as its cofactor.[10] The enzyme’s typical zinc content is 1.42 ug/mg.[11] The addition of cobalt or manganese ions cause the enzyme to take on different conformations, which suggests that the enzyme may be able to hydrolyze different dipeptides depending on which metal ions are present—aka the metal-content of one’s micronutrient intake could affect their renal dipeptidase’s ability to metabolize various dipeptides.[12]
Function
editThe primary function of DPEP1 is to hydrolyze various dipeptides in renal metabolism. Specifically, it has been found to hydrolyze glutathione and its conjugates such as leukotriene D (Kozak and Tate, 1982).
Several pieces of evidence suggest that DPEP1 is also responsible for the hydrolysis of the beta-lactam ring of various THM-class antibiotics, such as penem and carbapenem (Campbell et al., 1984). First, the metabolism of these THM-class antibiotics is known to be localized in the kidney, specifically by a membrane-bound protein. Second, the metabolism of these antibiotics is significantly hindered when the zinc concentration is altered, suggesting the enzyme responsible for the drugs’ metabolism is a zinc-metalloenzyme. Finally, when DPEP1 was experimentally added to penem and carbapenem antibiotics in vitro, the resulting products were structurally identical to their respective metabolites found in an organism's urine (8). The hydrolysis of these antibiotics hinders their antibacterial capabilities, so information on the specific structure of DPEPI is highly sought after in order to find viable inhibitors that could be taken along with these antibiotics to make them more effective.[13]
Earlier, beta-lactamase enzymes were thought to occur only in bacteria, where their probable function was in protecting the organisms against the action of beta-lactam antibiotics. These antibiotics exhibit selective toxicity against bacteria but virtual inertness against many eukaryotic cells (Adachi et al., 1990).[supplied by OMIM][14]
Reaction Mechanism
editWhen hydrolyzing a substrate, DPEP1 goes through a tetrahedral intermediate, after which the bridging solvent attacks the face of the carbonyl carbon of the scissile peptide bond.[15] Although DPEP1 shows preference for dipeptide substrates with D amino acids at the carboxy positions, it has been shown that DPEP1 can accommodate substrates with both D and L amino acids.[16]
Interactions
editCancer
editDPEP1 has been found to be highly expressed in colon tumor cells compared to normal colon cells—one study even found a ≥2 fold over-expression of DPEP1. Increased levels of DPEP1 have also been detected in colorectal cancer patients, suggesting that DPEP1 is a viable marker for disseminated colon tumor cells.[18]
References
edit- ^ a b c GRCh38: Ensembl release 89: ENSG00000015413 – Ensembl, May 2017
- ^ a b c GRCm38: Ensembl release 89: ENSMUSG00000019278 – Ensembl, May 2017
- ^ "Human PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- ^ "Mouse PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- ^ Armstrong, David J., Sunil K. Mukhopadhyay, and Benedict J. Campbell. "Physicochemical characterization of renal dipeptidase." Biochemistry 13.8 (1974): 1745-750. Web.
- ^ Keynan, Shoshana, Nicolette T. Habgood, Nigel M. Hooper, and Anthony J. Turner. "Site-Directed Mutagenesis of Conserved Cysteine Residues in Porcine Membrane Dipeptidase. Cys 361 Alone Is Involved in Disulfide-Linked Dimerization†." Biochemistry 35.38 (1996): 12511-2517. Web.
- ^ Nitanai, Yasushi, Yoshinori Satow, Hideki Adachi, and Masafumi Tsujimoto. "Crystal Structure of Human Renal Dipeptidase Involved in β-Lactam Hydrolysis." Journal of Molecular Biology 321.2 (2002): 177-84. Web.
- ^ Satoh, Susumu, Kazuyuki Ohtsuka, Yuriko Keida, Chihiro Kusunoki, Yoshiyuki Konta, Mineo Niwa, and Masanobu Kohsaka. "Gene structural analysis and expression of human renal dipeptidase." Biotechnology Progress 10.2 (1994): 134-40. Web.
- ^ Thoden, James B., Ricardo Marti-Arbona, Frank M. Raushel, and Hazel M. Holden. "High-Resolution X-Ray Structure of Isoaspartyl Dipeptidase from Escherichia coli†,‡." Biochemistry 42.17 (2003): 4874-882. Web.
- ^ Armstrong, David J., Sunil K. Mukhopadhyay, and Benedict J. Campbell. "Physicochemical characterization of renal dipeptidase." Biochemistry 13.8 (1974): 1745-750. Web.
- ^ Wu, Yong Qian, and Shahriar Mobashery. "Targeting renal dipeptidase (dehydropeptidase I) for inactivation by mechanism-based inactivators." Journal of Medicinal Chemistry 34.6 (1991): 1914-916. Web.
- ^ Hayman, Selma, Joselina S. Gatmaitan, and Elizabeth K. Patterson. "Relation of extrinsic and intrinsic metal ions to the specificity of a dipeptidase from Escherichia coli B." Biochemistry 13.22 (1974): 4486-494. Web.
- ^ Nitanai, Yasushi, Yoshinori Satow, Hideki Adachi, and Masafumi Tsujimoto. "Crystal Structure of Human Renal Dipeptidase Involved in β-Lactam Hydrolysis." Journal of Molecular Biology 321.2 (2002): 177-84. Web.
- ^ "Entrez Gene: DPEP1 dipeptidase 1 (renal)".
- ^ Thoden, James B., Ricardo Marti-Arbona, Frank M. Raushel, and Hazel M. Holden. "High-Resolution X-Ray Structure of Isoaspartyl Dipeptidase from Escherichia coli†,‡." Biochemistry 42.17 (2003): 4874-882. Web.
- ^ Wu, Yong Qian, and Shahriar Mobashery. "Targeting renal dipeptidase (dehydropeptidase I) for inactivation by mechanism-based inactivators." Journal of Medicinal Chemistry 34.6 (1991): 1914-916. Web.
- ^ Rual JF, Venkatesan K, Hao T, Hirozane-Kishikawa T, Dricot A, Li N, Berriz GF, Gibbons FD, Dreze M, Ayivi-Guedehoussou N, Klitgord N, Simon C, Boxem M, Milstein S, Rosenberg J, Goldberg DS, Zhang LV, Wong SL, Franklin G, Li S, Albala JS, Lim J, Fraughton C, Llamosas E, Cevik S, Bex C, Lamesch P, Sikorski RS, Vandenhaute J, Zoghbi HY, Smolyar A, Bosak S, Sequerra R, Doucette-Stamm L, Cusick ME, Hill DE, Roth FP, Vidal M (Oct 2005). "Towards a proteome-scale map of the human protein-protein interaction network". Nature. 437 (7062): 1173–8. Bibcode:2005Natur.437.1173R. doi:10.1038/nature04209. PMID 16189514. S2CID 4427026.
- ^ Mciver, C.m, J.m Lloyd, P.j Hewett, and J.e Hardingham. "Dipeptidase 1: a candidate tumor-specific molecular marker in colorectal carcinoma." Cancer Letters 209.1 (2004): 67-74. Web.
Further reading
edit- Hooper NM, Keen JN, Turner AJ (Jan 1990). "Characterization of the glycosyl-phosphatidylinositol-anchored human renal dipeptidase reveals that it is more extensively glycosylated than the pig enzyme". The Biochemical Journal. 265 (2): 429–33. doi:10.1042/bj2650429. PMC 1136904. PMID 2137335.
- Adachi H, Katayama T, Inuzuka C, Oikawa S, Tsujimoto M, Nakazato H (Sep 1990). "Identification of membrane anchoring site of human renal dipeptidase and construction and expression of a cDNA for its secretory form". The Journal of Biological Chemistry. 265 (25): 15341–5. doi:10.1016/S0021-9258(18)77261-X. PMID 2168407.
- Adachi H, Tawaragi Y, Inuzuka C, Kubota I, Tsujimoto M, Nishihara T, Nakazato H (Mar 1990). "Primary structure of human microsomal dipeptidase deduced from molecular cloning". The Journal of Biological Chemistry. 265 (7): 3992–5. doi:10.1016/S0021-9258(19)39692-9. PMID 2303490.
- Adachi H, Kubota I, Okamura N, Iwata H, Tsujimoto M, Nakazato H, Nishihara T, Noguchi T (Jun 1989). "Purification and characterization of human microsomal dipeptidase". Journal of Biochemistry. 105 (6): 957–61. doi:10.1093/oxfordjournals.jbchem.a122787. PMID 2768222.
- Austruy E, Jeanpierre C, Antignac C, Whitmore SA, Van Cong N, Bernheim A, Callen DF, Junien C (Mar 1993). "Physical and genetic mapping of the dipeptidase gene DPEP1 to 16q24.3". Genomics. 15 (3): 684–7. doi:10.1006/geno.1993.1126. PMID 7682195.
- Satoh S, Ohtsuka K, Keida Y, Kusunoki C, Konta Y, Niwa M, Kohsaka M (1994). "Gene structural analysis and expression of human renal dipeptidase". Biotechnology Progress. 10 (2): 134–40. doi:10.1021/bp00026a002. PMID 7764673. S2CID 34807766.
- Adachi H, Katayama T, Nakazato H, Tsujimoto M (Apr 1993). "Importance of Glu-125 in the catalytic activity of human renal dipeptidase". Biochimica et Biophysica Acta (BBA) - Protein Structure and Molecular Enzymology. 1163 (1): 42–8. doi:10.1016/0167-4838(93)90276-w. PMID 8097406.
- Satoh S, Kusunoki C, Konta Y, Niwa M, Kohsaka M (Feb 1993). "Cloning and structural analysis of genomic DNA for human renal dipeptidase". Biochimica et Biophysica Acta (BBA) - Gene Structure and Expression. 1172 (1–2): 181–3. doi:10.1016/0167-4781(93)90289-p. PMID 8439558.
- Satoh S, Keida Y, Konta Y, Maeda M, Matsumoto Y, Niwa M, Kohsaka M (Jun 1993). "Purification and molecular cloning of mouse renal dipeptidase". Biochimica et Biophysica Acta (BBA) - Protein Structure and Molecular Enzymology. 1163 (3): 234–42. doi:10.1016/0167-4838(93)90157-m. PMID 8507661.
- Kera Y, Liu Z, Matsumoto T, Sorimachi Y, Nagasaki H, Yamada RH (May 1999). "Rat and human membrane dipeptidase: tissue distribution and developmental changes". Comparative Biochemistry and Physiology B. 123 (1): 53–8. doi:10.1016/S0305-0491(99)00039-5. PMID 10425712.
- Nitanai Y, Satow Y, Adachi H, Tsujimoto M (Aug 2002). "Crystal structure of human renal dipeptidase involved in beta-lactam hydrolysis". Journal of Molecular Biology. 321 (2): 177–84. doi:10.1016/S0022-2836(02)00632-0. PMID 12144777.
- McIver CM, Lloyd JM, Hewett PJ, Hardingham JE (Jun 2004). "Dipeptidase 1: a candidate tumor-specific molecular marker in colorectal carcinoma". Cancer Letters. 209 (1): 67–74. doi:10.1016/j.canlet.2003.11.033. PMID 15145522.
- Zhang Z, Henzel WJ (Oct 2004). "Signal peptide prediction based on analysis of experimentally verified cleavage sites". Protein Science. 13 (10): 2819–24. doi:10.1110/ps.04682504. PMC 2286551. PMID 15340161.
- Rual JF, Venkatesan K, Hao T, Hirozane-Kishikawa T, Dricot A, Li N, Berriz GF, Gibbons FD, Dreze M, Ayivi-Guedehoussou N, Klitgord N, Simon C, Boxem M, Milstein S, Rosenberg J, Goldberg DS, Zhang LV, Wong SL, Franklin G, Li S, Albala JS, Lim J, Fraughton C, Llamosas E, Cevik S, Bex C, Lamesch P, Sikorski RS, Vandenhaute J, Zoghbi HY, Smolyar A, Bosak S, Sequerra R, Doucette-Stamm L, Cusick ME, Hill DE, Roth FP, Vidal M (Oct 2005). "Towards a proteome-scale map of the human protein-protein interaction network". Nature. 437 (7062): 1173–8. Bibcode:2005Natur.437.1173R. doi:10.1038/nature04209. PMID 16189514. S2CID 4427026.