Haplogroup M, AKA M-P256 and Haplogroup K2b1b (previously K2b1d) is a Y-chromosome DNA haplogroup. M-P256 is a descendant haplogroup of Haplogroup K2b1, and is believed to have first appeared between 32,000 to 47,000 years ago.[1]
| Haplogroup M-P256 | |
|---|---|
 | |
| Possible time of origin | 32,000-47,000 years BP[1] |
| Possible place of origin | Wallacea (eastern Indonesia) or New Guinea [2] |
| Ancestor | K2b1 |
| Defining mutations | P256 |
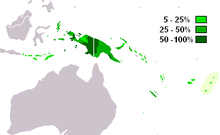
M-P256 is the most frequently occurring Y-chromosome haplogroup in West Papua and western Papua New Guinea.[3] In addition, M-P256 is also found in neighboring parts of Melanesia, Indonesia and indigenous Australians.
Phylogenetic structure
editThis phylogenetic tree of haplogroup subclades is based primarily on the trees published by YCC in 2008[4] and ISOGG in 2016.[5]
- M* (P256)
- M1 (M4, M5/P73, M106, M186, M189, M296, P35)
- M1a(P34_1, P34_2, P34_3, P34_4, P34_5)
- M1a1 (P51)
- M1a2 (P94)
- M1b (P87)
- M1b1 (M104_1/P22_1, M104_2/P22_2)
- M1b1a (M16)
- M1b1b (M83)
- M1b1 (M104_1/P22_1, M104_2/P22_2)
- M1a(P34_1, P34_2, P34_3, P34_4, P34_5)
- M2 (M353, M387)
- M2a (M177/SRY9138)
- M3 (P117, P118)
- M1 (M4, M5/P73, M106, M186, M189, M296, P35)
Distribution
editM* (M-P256*)
editThe paragroup M-P256* is found at a low frequency in New Guinea (6.3%) and Flores (2.5%).[2]
M1 (M-M4)
edit| Haplogroup M-M4 | |
|---|---|
| Possible time of origin | 8,200 [3,800–20,600] years BP[3] |
| Possible place of origin | Southeast Asia - Melanesia[citation needed] |
| Ancestor | M-P256 |
| Defining mutations | M4, M5/P73, M106, M186, M189, M296, P35[citation needed] |
This group is found frequently in New Guinea and Melanesia, with a moderate distribution in neighboring parts of Indonesia, Micronesia, and Polynesia.
- Una 100%[3]
- Ketengban 100%[3]
- Awyu 100%[3]
- Citak 86%[3]
- Asmat 75%[3]
- West Papua
- Kombai/Korowai 46%[3]
- Papua New Guinea
- Tolai (New Britain) 31%[3]
- Trobriand Islands 30%[3]
- Maluku (Moluccas) 21%[3]
- Torres Strait Islanders (Australia): up to 2.0% – i.e. 0.9% of samples, when 45% of the total were deemed to be "non-indigenous"[6]
An extreme geographical outlier was apparently identified in a 2012 study, which reported a Hazara individual from Mazar-e Sharif, Afghanistan, with M1 (among a sample of 60 males from the mentioned area).[7] The Hazara individual carried the SNP M186 (which is believed to be equivalent to M4).
| Old names (YCC 2002/2008) | M-M4 |
| Jobling and Tyler-Smith 2000 | 24 |
| Underhill 2000 | VIII |
| Hammer 2001 | 1U |
| Karafet 2001 | 37 |
| Semino 2000 | Eu16 |
| Su 1999 | H17 |
| Capelli 2001 | E |
| YCC 2002 (Longhand) | M* |
| YCC 2005 (Longhand) | M |
| YCC 2008 (Longhand) | M1 |
| YCC 2010r (Longhand) | M1 |
M1a (M-P34)
editM1a (M-P34) is the most frequently occurring Y-chromosome DNA haplogroup in Western New Guinea. It is also found with moderate frequency in neighboring parts of Indonesia (Maluku, Nusa Tenggara) and throughout Papua New Guinea, including offshore islands.[3][8]
| Old names (YCC 2002/2008) | M-P34 |
| Jobling and Tyler-Smith 2000 | 24 |
| Underhill 2000 | VIII |
| Hammer 2001 | 1U |
| Karafet 2001 | 37 |
| Semino 2000 | Eu16 |
| Su 1999 | H17 |
| Capelli 2001 | E |
| YCC 2002 (Longhand) | M1 |
| YCC 2005 (Longhand) | M1 |
| YCC 2008 (Longhand) | M1a |
| YCC 2010r (Longhand) | M1a |
M1b (M-P87)
editM1b M-P87(xM104/P22) has been found in approximately 18% (20/109) of a pool of samples from New Ireland, approximately 12% (5/43) of a sample of Lavongai from New Hanover, approximately 5% (19/395) of a pool of samples from New Britain (and, in particular, in about 24% (15/63) of Baining from East New Britain), in addition to one Saposa individual from northern Bougainville, and another individual from the north coast of Papua New Guinea.[1]
The subclade M1b1 (M104_1/P22_1, M104_2/P22_2) is found frequently in populations of the Bismarck Archipelago and Bougainville Island, with a moderate distribution in New Guinea, Fiji, Tonga, East Futuna, and Samoa.[1][9]
| Old names (YCC 2002/2008) | M-P22 |
| Jobling and Tyler-Smith 2000 | 24 |
| Underhill 2000 | VIII |
| Hammer 2001 | 1U |
| Karafet 2001 | 38 |
| Semino 2000 | Eu16 |
| Su 1999 | H17 |
| Capelli 2001 | E |
| YCC 2002 (Longhand) | M2* |
| YCC 2005 (Longhand) | M2a |
| YCC 2008 (Longhand) | M1b1 |
| YCC 2010r (Longhand) | M1b1 |
M2 (M-M353)
editM2 is found at a low frequency in Fiji and East Futuna.[10]
The subclade M2a (M-M177, also referred to as M-SRY9138) has been found in one Nasioi individual from the eastern coast of Bougainville and in one individual from Malaita Province of the Solomon Islands.[11]
Alternative names previously used within peer-reviewed literature for the M2a subclade are listed below.
| Old names (YCC 2002/2008) | K-SRY9138/M-SRY9138 AKA M-M177 |
| Jobling and Tyler-Smith 2000 | 23 |
| Underhill 2000 | VIII |
| Hammer 2001 | 1E |
| Karafet 2001 | 25 |
| Semino 2000 | Eu16 |
| Su 1999 | H5 |
| Capelli 2001 | F |
| YCC 2002 (Longhand) | K1 |
| YCC 2005 (Longhand) | K1 |
| YCC 2008 (Longhand) | M2a |
| YCC 2010r (Longhand) | M2a |
M3 (M-P117)
editM3 (P117, P118) is found frequently in populations of New Britain, and is also observed occasionally in northern Bougainville, Fiji, and East Futuna.[9][1]
Previous phylogenetic history
editPrior to 2002, at least seven different naming systems for the Y chromosome phylogenetic tree were used within academic literature, leading to considerable confusion. To resolve this, in 2002, major research groups collaborated to form the Y-Chromosome Consortium (YCC). This resulted in a joint paper publication that contained a single new tree as a standard to use by the scientific community. Later, another group of scientists with an interest in population genetics and genetic genealogy worked to continually improve the naming system.
The table below brings together the nomenclature used in Haplogroup M studies, prior to the landmark 2002 YCC Tree, enabling researchers reviewing older literature to quickly convert between the different nomenclatures that were in use.
| YCC 2002/2008 (Shorthand) | (α) | (β) | (γ) | (δ) | (ε) | (ζ) | (η) | YCC 2002 (Longhand) | YCC 2005 (Longhand) | YCC 2008 (Longhand) | YCC 2010r (Longhand) | ISOGG 2006 | ISOGG 2007 | ISOGG 2008 | ISOGG 2009 | ISOGG 2010 | ISOGG 2011 | ISOGG 2012 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| M4 | 24 | VIII | 1U | 37 | Eu16 | H17 | E | M* | M | M1 | M1 | - | - | - | - | - | - | - |
| M-P34 | 24 | VIII | 1U | 37 | Eu16 | H17 | E | M1 | M1 | M1a | M1a | - | - | - | - | - | - | - |
| M-P22/M-M104 | 24 | VIII | 1U | 38 | Eu16 | H17 | E | M2* | M2a | M1b1 | M1b1 | - | - | - | - | - | - | - |
| M-M16 | 24 | VIII | 1U | 39 | Eu16 | H17 | E | M2a | M2a1 | M1b1a | M1b1a | - | - | - | - | - | - | - |
| M-M83 | 24 | VIII | 1U | 38 | Eu16 | H17 | E | M2b | M2a2 | M1b1b | M1b1b | - | - | - | - | - | - | - |
| K-SRY9138/M-SRY9138 | 23 | VIII | 1E | 25 | Eu16 | H5 | F | K1 | K1 | M2a | M2a | - | - | - | - | - | - | - |
- Sources
The following research teams per their publications were represented in the creation of the YCC Tree.
Karafet's 2008 paper introduced a number of changes, compared to the previous 2006 ISOGG tree.[4] Before the discovery of the P256 marker, the current subgroup M-M4 (defined by the M4 marker) previously represented the whole of Haplogroup M-P256; and subgroups M2 and M3 were formerly classed as subgroups K1 and K7 of the parent Haplogroup K.[citation needed]
References
edit- ^ a b c d e Scheinfeldt, Laura; Friedlaender, Françoise; Friedlaender, Jonathan; Latham, Krista; Koki, George; Karafet, Tatyana; Hammer, Michael; Lorenz, Joseph (2006-06-05). "Unexpected NRY Chromosome Variation in Northern Island Melanesia". Molecular Biology and Evolution. 23 (8): 1628–1641. doi:10.1093/molbev/msl028. ISSN 1537-1719. PMID 16754639.
- ^ a b Tatiana M. Karafet, Brian Hallmark, Murray P. Cox, Herawati Sudoyo, Sean Downey, J. Stephen Lansing and Michael F. Hammer, "Major East–West Division Underlies Y Chromosome Stratification across Indonesia", Molecular Biological Evolution, (2010), vol. 27, no. 8, pp. 1833-1844.
- ^ a b c d e f g h i j k l m n o p Kayser, Manfred; Brauer, Silke; Weiss, Gunter; Schiefenhövel, Wulf; Underhill, Peter; Shen, Peidong; Oefner, Peter; Tommaseo-Ponzetta, Mila; Stoneking, Mark (2003). "Reduced Y-Chromosome, but Not Mitochondrial DNA, Diversity in Human Populations from West New Guinea". The American Journal of Human Genetics. 72 (2): 281–302. doi:10.1086/346065. ISSN 0002-9297. PMC 379223. PMID 12532283.
- ^ a b Karafet, Tatiana M.; Mendez, Fernando L.; Meilerman, Monica B.; Underhill, Peter A.; Zegura, Stephen L.; Hammer, Michael F. (2008-05-01). "New binary polymorphisms reshape and increase resolution of the human Y chromosomal haplogroup tree". Genome Research. 18 (5): 830–838. doi:10.1101/gr.7172008. ISSN 1088-9051. PMC 2336805. PMID 18385274.
- ^ "ISOGG 2018 Y-DNA Haplogroup M". isogg.org. Retrieved 2024-09-18.
- ^ Nagle, Nano; Ballantyne, Kaye N.; van Oven, Mannis; Tyler-Smith, Chris; Xue, Yali; Taylor, Duncan; Wilcox, Stephen; Wilcox, Leah; Turkalov, Rust; van Oorschot, Roland A.H.; McAllister, Peter; Williams, Lesley; Kayser, Manfred; Mitchell, Robert J.; The Genographic Consortium (2016). "Antiquity and diversity of aboriginal Australian Y -chromosomes". American Journal of Physical Anthropology. 159 (3): 367–381. doi:10.1002/ajpa.22886. ISSN 0002-9483. PMID 26515539.
- ^ Haber, Marc; Platt, Daniel E.; Bonab, Maziar Ashrafian; Youhanna, Sonia C.; Soria-Hernanz, David F.; Martínez-Cruz, Begoña; Douaihy, Bouchra; Ghassibe-Sabbagh, Michella; Rafatpanah, Hoshang; Ghanbari, Mohsen; Whale, John; Balanovsky, Oleg; Wells, R. Spencer; Comas, David; Tyler-Smith, Chris (2012-03-28). "Afghanistan's Ethnic Groups Share a Y-Chromosomal Heritage Structured by Historical Events". PLOS ONE. 7 (3): e34288. Bibcode:2012PLoSO...734288H. doi:10.1371/journal.pone.0034288. ISSN 1932-6203. PMC 3314501. PMID 22470552.
- ^ Karafet, Tatiana M; Lansing, J. S; Redd, Alan J; Watkins, Joseph C; Surata, S. P. K; Arthawiguna, W. A; Mayer, Laura; Bamshad, Michael; Jorde, Lynn B; Hammer, Michael F (2005). "Balinese Y-Chromosome Perspective on the Peopling of Indonesia: Genetic Contributions from Pre-Neolithic Hunter-Gatherers, Austronesian Farmers, and Indian Traders". Human Biology. 77 (1): 93–114. doi:10.1353/hub.2005.0030. ISSN 1534-6617. PMID 16114819.
- ^ a b Kayser, M.; Choi, Y.; van Oven, M.; Mona, S.; Brauer, S.; Trent, R. J.; Suarkia, D.; Schiefenhovel, W.; Stoneking, M. (2008-04-03). "The Impact of the Austronesian Expansion: Evidence from mtDNA and Y Chromosome Diversity in the Admiralty Islands of Melanesia". Molecular Biology and Evolution. 25 (7): 1362–1374. doi:10.1093/molbev/msn078. ISSN 0737-4038.
- ^ Kayser, Manfred; Brauer, Silke; Cordaux, Richard; Casto, Amanda; Lao, Oscar; Zhivotovsky, Lev A.; Moyse-Faurie, Claire; Rutledge, Robb B.; Schiefenhoevel, Wulf; Gil, David; Lin, Alice A.; Underhill, Peter A.; Oefner, Peter J.; Trent, Ronald J.; Stoneking, Mark (2006). "Melanesian and Asian Origins of Polynesians: mtDNA and Y Chromosome Gradients Across the Pacific". Molecular Biology and Evolution. 23 (11): 2234–2244. doi:10.1093/molbev/msl093. hdl:11858/00-001M-0000-0010-0145-0. ISSN 1537-1719. PMID 16923821.
- ^ Cox, Murray P.; Mirazón Lahr, Marta (2006). "Y-chromosome diversity is inversely associated with language affiliation in paired Austronesian- and Papuan-speaking communities from Solomon Islands". American Journal of Human Biology. 18 (1): 35–50. doi:10.1002/ajhb.20459. ISSN 1042-0533. PMID 16378340.
Sources for conversion tables
edit- Capelli, Cristian; Wilson, James F.; Richards, Martin; Stumpf, Michael P.H.; et al. (February 2001). "A Predominantly Indigenous Paternal Heritage for the Austronesian-Speaking Peoples of Insular Southeast Asia and Oceania". The American Journal of Human Genetics. 68 (2): 432–443. doi:10.1086/318205. PMC 1235276. PMID 11170891.
- Hammer, Michael F.; Karafet, Tatiana M.; Redd, Alan J.; Jarjanazi, Hamdi; et al. (1 July 2001). "Hierarchical Patterns of Global Human Y-Chromosome Diversity". Molecular Biology and Evolution. 18 (7): 1189–1203. doi:10.1093/oxfordjournals.molbev.a003906. PMID 11420360.
- Jobling, Mark A.; Tyler-Smith, Chris (2000), "New uses for new haplotypes", Trends in Genetics, 16 (8): 356–62, doi:10.1016/S0168-9525(00)02057-6, PMID 10904265
- Kaladjieva, Luba; Calafell, Francesc; Jobling, Mark A; Angelicheva, Dora; et al. (February 2001). "Patterns of inter- and intra-group genetic diversity in the Vlax Roma as revealed by Y chromosome and mitochondrial DNA lineages". European Journal of Human Genetics. 9 (2): 97–104. doi:10.1038/sj.ejhg.5200597. PMID 11313742.
- Karafet, Tatiana; Xu, Liping; Du, Ruofu; Wang, William; et al. (September 2001). "Paternal Population History of East Asia: Sources, Patterns, and Microevolutionary Processes". The American Journal of Human Genetics. 69 (3): 615–628. doi:10.1086/323299. PMC 1235490. PMID 11481588.
- Semino, O.; Passarino, G; Oefner, PJ; Lin, AA; et al. (2000), "The Genetic Legacy of Paleolithic Homo sapiens sapiens in Extant Europeans: A Y Chromosome Perspective", Science, 290 (5494): 1155–9, Bibcode:2000Sci...290.1155S, doi:10.1126/science.290.5494.1155, PMID 11073453
- Su, Bing; Xiao, Junhua; Underhill, Peter; Deka, Ranjan; et al. (December 1999). "Y-Chromosome Evidence for a Northward Migration of Modern Humans into Eastern Asia during the Last Ice Age". The American Journal of Human Genetics. 65 (6): 1718–1724. doi:10.1086/302680. PMC 1288383. PMID 10577926.
- Underhill, Peter A.; Shen, Peidong; Lin, Alice A.; Jin, Li; et al. (November 2000). "Y chromosome sequence variation and the history of human populations". Nature Genetics. 26 (3): 358–361. doi:10.1038/81685. PMID 11062480.