This article may be too technical for most readers to understand. (May 2017) |
HLA-A3 (A3) is a human leukocyte antigen serotype within HLA-A serotype group. The serotype is determined by the antibody recognition of α3 subset of HLA-A α-chains. For A3, the alpha, "A", chain are encoded by the HLA-A*03 allele group and the β-chain are encoded by B2M locus.[1] This group currently is dominated by A*03:01. A3 and A*03 are almost synonymous in meaning. A3 is more common in Europe, it is part of the longest known multigene haplotype, A3~B7~DR15~DQ6.[2]
| HLA-A3 | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| (MHC Class I, A cell surface antigen) | ||||||||||
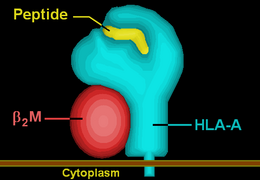 HLA-A3 | ||||||||||
| About | ||||||||||
| Protein | transmembrane receptor/ligand | |||||||||
| Structure | αβ heterodimer | |||||||||
| Subunits | HLA-A*03--, β2-microglobulin | |||||||||
| Older names | HL-A3 | |||||||||
| Subtypes | ||||||||||
| ||||||||||
| Rare alleles | ||||||||||
| ||||||||||
| Alleles link-out to IMGT/HLA database at EBI | ||||||||||
Serotype edit
| A*03 | A3 | Sample |
| allele | % | size (N) |
| *03:01 | 99 | 3504 |
| *03:02 | 78 | 342 |
| *03:05 | 20 | 5 |
A3 is primarily composed of A*03:01 and *03:02 which serotype well with anti-A3 antibodies. There are 26 non-synonymous variants of A*03, 4 nulls, and 22 protein variants.
Associated diseases edit
A3 serotype is a secondary risk factor for myasthenia gravis[4] and lower CD8+ levels in hemochromatosis patients.[5][6] The HFE (Hemochromatosis) locus lies between A3 and B7 within the A3~DQ6 superhaplotype.[7]
In HIV edit
HLA-A3 selects HIV evolution for a mutation Gag KK9 epitope and results in a rapid decline in the CD8 T-cell response. CD8 T-cells are responsible for quickly killing HIV infected CD4+ cells.[8] This type of evolved response may not be specific for HLA-A3 and since HIV is capable of adapting quickly in situ to selective factors.
Alleles edit
| Study population | Freq. (in %)[9] |
|---|---|
| Finland | 25.0 |
| India Tamil Nadu Nadar | 20.5 |
| Czech Republic | 18.9 |
| Belgium | 17.1 |
| Ireland Northern | 14.3 |
| Australia New South Wales | 13.8 |
| Georgia Svaneti Svans | 13.8 |
| Georgia Tbilisi Georgian… | 13.8 |
| Pakistan Burusho | 13.0 |
| Cape Verde Northwestern I… | 12.0 |
| Ireland South | 11.6 |
| Romanian | 11.6 |
| Croatia | 11.0 |
| USA Caucasians (3) | 10.8 |
| Italy North (1) | 9.7 |
| USA African Americans (2) | 9.4 |
| India Mumbai Marathas | 9.3 |
| India West Bhils | 9.0 |
| Portugal Centre | 9.0 |
| Pakistan Pathan | 8.7 |
| Cameroon Beti | 8.6 |
| Cameroon Bamileke | 7.8 |
| Pakistan Kalash | 7.5 |
| Saudi Arabia Guraiat and … | 7.5 |
| USA Hispanic | 7.3 |
| India North Delhi | 7.2 |
| Madeira | 7.0 |
| Russia Tuva (2) | 6.9 |
| Tunisia | 6.7 |
| USA North American Native… | 6.7 |
| Cape Verde Southeastern I… | 6.5 |
| Azores Santa Maria and Sa… | 6.4 |
| Pakistan Baloch | 6.3 |
| Guinea Bissau | 6.2 |
| Israel Arab Druse | 6.0 |
| South African Natal Zulu | 6.0 |
| USA South Texas Hispanics | 6.0 |
| India North Hindus | 5.8 |
| Zambia Lusaka | 5.8 |
| Iran Baloch | 5.6 |
| Uganda Kampala | 5.5 |
| Jordan Amman | 5.2 |
| Kenya | 5.2 |
| Brazil | 5.1 |
| Oman | 5.1 |
| Cameroon Pygmy Baka | 5.0 |
| Georgia Tbilisi Kurds | 5.0 |
| Sudanese | 4.8 |
| Bulgaria | 4.6 |
| Pakistan Brahui | 4.6 |
| China Beijing | 4.5 |
| Australia Indig. Cape Yor… | 4.4 |
| China Inner Mongolia | 4.4 |
| Mali Bandiagara | 4.4 |
| Pakistan Sindhi | 4.4 |
| China Qinghai Hui | 4.1 |
| Zimbabwe Harare Shona | 4.0 |
| Cameroon Sawa | 3.8 |
| Senegal Niokholo Mandenka | 3.8 |
| Kenya Luo | 3.6 |
| Morocco Nador Metalsa Cla… | 3.4 |
| Kenya Nandi | 3.1 |
| Mex. Guadalajara Mestizos… | 3.1 |
| China Guangzhou | 2.9 |
| China North Han | 2.9 |
| Mongolia Buriat | 2.9 |
| China Tibetans | 2.5 |
| China Yunnan Nu | 2.5 |
| Mexico Mestizos | 2.4 |
| American Samoa | 2.0 |
| Singapore Javanese Indone… | 2.0 |
| Allele frequencies presented, only | |
| Study population | Freq. (in %)[9] |
|---|---|
| Georgia Tbilisi Kurds | 8.3 |
| Morocco Nador Metalsa Cla… | 4.1 |
| Pakistan Karachi Parsi | 2.8 |
| Israel Arab Druse | 2.5 |
| India New Delhi | 2.3 |
| Sudanese | 2.3 |
| India North Delhi | 2.2 |
| India West Coast Parsis | 2.0 |
| Portugal Centre | 2.0 |
| India Andhra Pradesh Goll… | 1.7 |
| Pakistan Kalash | 1.7 |
| Pakistan Baloch | 1.6 |
| Georgia Tbilisi Georgian… | 1.4 |
| Saudi Arabia Guraiat and … | 1.4 |
| Oman | 1.3 |
| Pakistan Sindhi | 1.3 |
| India North Hindus | 1.0 |
| Pakistan Pathan | 1.0 |
| South Africa Natal Tamil | 1.0 |
| USA South Texas Hispanics | 0.8 |
| Allele frequencies presented, only | |
Associated diseases edit
A*03:01 modulates increased risk for multiple sclerosis[10]
A3~B haplotypes edit
A3-B7 is part of the A3~DQ2 superhaplotype
A3-B8 (Romania, svanS)
A3~B35 (Bulgaria, Croatia, E. Black Sea)
A3~B55 (E. Black Sea)
A3~Cw7~B7 edit
A3~B7 is bimodal in frequency in Europe with one node in Ireland and the other in Switzerland, relatively speaking Switzerland appears to be higher. A3~Cw7~B7 is one of the most common multigene haplotypes in the western world, particularly in Central and Eastern Europe.
A*03:01 ~ C*07:02 ~ B*07:02 ~ DRB1*15:01 ~ DQA1*01:02 ~ DQB1*06:02
References edit
- ^ Arce-Gomez B, Jones EA, Barnstable CJ, Solomon E, Bodmer WF (February 1978). "The genetic control of HLA-A and B antigens in somatic cell hybrids: requirement for beta2 microglobulin". Tissue Antigens. 11 (2): 96–112. doi:10.1111/j.1399-0039.1978.tb01233.x. PMID 77067.
- ^ Horton R, Gibson R, Coggill P, et al. (January 2008). "Variation analysis and gene annotation of eight MHC haplotypes: the MHC Haplotype Project". Immunogenetics. 60 (1): 1–18. doi:10.1007/s00251-007-0262-2. PMC 2206249. PMID 18193213.
- ^ Allele Query Form IMGT/HLA - European Bioinformatics Institute
- ^ Machens A, Löliger C, Pichlmeier U, Emskötter T, Busch C, Izbicki J (1999). "Correlation of thymic pathology with HLA in myasthenia gravis". Clinical Immunology. 91 (3): 296–301. doi:10.1006/clim.1999.4710. PMID 10370374.
- ^ Barton J, Wiener H, Acton R, Go R (2005). "HLA haplotype A*03-B*07 in hemochromatosis probands with HFE C282Y homozygosity: frequency disparity in men and women and lack of association with severity of iron overload". Blood Cells Mol Dis. 34 (1): 38–47. doi:10.1016/j.bcmd.2004.08.022. PMID 15607698.
- ^ Cruz E, Vieira J, Almeida S, Lacerda R, Gartner A, Cardoso C, Alves H, Porto G (2006). "A study of 82 extended HLA haplotypes in HFE-C282Y homozygous hemochromatosis subjects: relationship to the genetic control of CD8+ T-lymphocyte numbers and severity of iron overload". BMC Med Genet. 7: 16. doi:10.1186/1471-2350-7-16. PMC 1413516. PMID 16509978.
- ^ Olsson KS, Ritter B, Hansson N, Chowdhury RR (July 2008). "HLA haplotype map of river valley populations with hemochromatosis traced through five centuries in Central Sweden". Eur. J. Haematol. 81 (1): 36–46. doi:10.1111/j.1600-0609.2008.01078.x. PMID 18363869. S2CID 30689398.
- ^ Allen TM, Altfeld M, Yu XG, et al. (July 2004). "Selection, transmission, and reversion of an antigen-processing cytotoxic T-lymphocyte escape mutation in human immunodeficiency virus type 1 infection". J. Virol. 78 (13): 7069–78. doi:10.1128/JVI.78.13.7069-7078.2004. PMC 421658. PMID 15194783.
- ^ a b Middleton, D.; Menchaca, L.; Rood, H.; Komerofsky, R. (2003). "New allele frequency database: http://www.allelefrequencies.net". Tissue Antigens. 61 (5): 403–407. doi:10.1034/j.1399-0039.2003.00062.x. PMID 12753660.
- ^ Fogdell-Hahn A, Ligers A, Grønning M, Hillert J, Olerup O (2000). "Multiple sclerosis: a modifying influence of HLA class I genes in an HLA class II associated autoimmune disease". Tissue Antigens. 55 (2): 140–8. doi:10.1034/j.1399-0039.2000.550205.x. PMID 10746785.