The SymE-SymR toxin-antitoxin system consists of a small symbiotic endonuclease toxin, SymE, and a non-coding RNA symbiotic RNA antitoxin, SymR, which inhibits SymE translation.[1] SymE-SymR is a type I toxin-antitoxin system, and is under regulation by the antitoxin, SymR.[2] The SymE-SymR complex is believed to play an important role in recycling damaged RNA and DNA.[1] The relationship and corresponding structures of SymE and SymR provide insight into the mechanism of toxicity and overall role in prokaryotic systems.
| SymR | |
|---|---|
 Conserved secondary structure of SymR RNA. | |
| Identifiers | |
| Symbol | SymR |
| Rfam | RF01809 |
| Other data | |
| RNA type | Antisense RNA |
| Domain(s) | E. coli |
| PDB structures | PDBe |
| SymE Toxin of Type I toxin-antitoxin system | |||||||||
|---|---|---|---|---|---|---|---|---|---|
 SymE Toxin of Type I toxin-antitoxin system | |||||||||
| Identifiers | |||||||||
| Symbol | SymE_toxin | ||||||||
| Pfam | PF13957 | ||||||||
| InterPro | IPR020883 | ||||||||
| PROSITE | PS51740 | ||||||||
| |||||||||
| https://swissmodel.expasy.org/repository/uniprot/P39394 | |||||||||
Discovery
editSymR was originally labelled RyjC and is a 77 nucleotide (nt) RNA with a σ70 promoter. RyjC was found to overlap the yjiW open reading frame on the opposite strand by 6 nt, and was characterized as an antisense RNA which bound the 5' untranslated region of yjiW.[3] Further study led to the renaming of both yjiW and RyjC to SymE (SOS-induced yjiW gene with similarity to MazE) and SymR, respectively.[1] Despite similarities to the AbrB superfamily, the SymE family has been exclusively found in proteobacteria.[1]
Relationship between SymE and SymR
editThe SymR antisense RNA is transcribed 3 nt behind the SymE start codon which is why the SymR promoter is considered embedded within the SymE codon.[2] As a result, SymR blocks RNA translation of SymE by antisense binding, suggesting that this ultimately leads to SymR mRNA degradation.[4] Amino acid analysis has concluded that SymE may have evolved into an RNA cleavage protein that exhibits toxin-like behavior due to transcription factors or antitoxins.[2] In contrast to other common toxin-antitoxin systems, the SymR antitoxin is more stable than the SymE toxin.[1]
Following DNA damage, the SOS response represses transcription of SymR RNA, allowing SymE toxin to degrade potentially damaged mRNA until DNA has been repaired.[1] Conversely, SymE is tightly repressed by LexA repressor binding sites, SymR, and the Lon protease.[2] These three factors are present at multiple levels where LexA is involved in transcription downregulation, SymR RNA is involved in translation downregulation, and Lon protease is involved in protein degradation.[1][2] The extent of repression on SymE is dependent on the additive power of LexA, SymR, and Lon protease.[2] Overall, SymE synthesis is slow since its activity is highly dependent on DNA repair proteins.[2] In the cellular environment, mitomycin C damages DNA which leads to an overexpression of SymE mRNA to initiate DNA repair.[5]
Toxicity
editThe overexpression of SymE demonstrated negative effects on the growth of colony-forming cells when tested in vitro.[1] SymE exhibits its toxicity by repressing global translation within the cell, cleaving mRNA in a similar manner to MazF, another toxin.[6] Quantitative Northern blot experiments showed that SymR RNA is present in cells at 10 times the concentration of SymE mRNA (0.02 fmol μg−1 and 0.2 fmol μg−1).[1]
Structure
editSymE
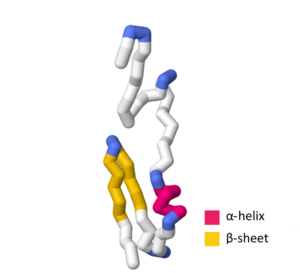
editThe SymE toxin consists of 113 amino acids.[5] When evaluating the amino acid sequence and tertiary structure of SymE, strong similarities were found which resemble the AbrB superfamily.[1] This superfamily mainly functions as transcription factors or antitoxins; however, the similarity of SymE to the primary sequence and tertiary structure of the AbrB superfamily suggests that SymE proteins experienced an evolutionary shift from a transcription factor or antitoxin to a RNA-associating protein that exhibits toxin behavior.[1] Between the AbrB superfamily protein structure and the SymE protein structure, there are several key hydrophobic residues that are highly conserved in the -helix at the center of the protein as well as the strand-1.[1] Despite these key similarities, SymE exhibits polar residues not found in the general structure of the AbrB superfamily, indicating that these residues may have a role in the SymE RNA cleavage ability.[1]
SWISS-MODEL contains more than several experimental structures and theoretical homology models that define certain aspects of the SymE primary sequence and tertiary structure. The UniProtKB accession number P39394 indicates the general structure of the SymE toxin in Escherichia coli (strain K12).[1][7] In the SWISS-MODEL SymE theoretical model, the -helix contains amino acids G44, Q45, W46, L47, E48, A49, and A50.[8][9][10][11][12] The strand-1 contains amino acids G55, T56, A57, V58, D59, V60, K61, V62, I67, V68, L69, T70, A71, Q72, P73, and P74 with the -turn containing M63, E64, G65, and C66.[8][9][10][11][12]
SymR
editSymR is an antisense RNA meaning its secondary structure has characteristic stem-and-loop elements as well as unpaired regions flanking the structure.[13] The predicted secondary structure of SymR showcases a loop containing the nucleotide sequence CCAG.[4] This characteristic loop is shared with the lstR-1 and OhsC RNA proteins and is predicted to be a binding site for other proteins.[4] Currently, there are no known files on the RCSB protein data bank or SWISS-MODEL repository that indicate a predicted tertiary structure of SymR.
See also
editReferences
edit- ^ a b c d e f g h i j k l m n Kawano M, Aravind L, Storz G (May 2007). "An antisense RNA controls synthesis of an SOS-induced toxin evolved from an antitoxin". Molecular Microbiology. 64 (3): 738–54. doi:10.1111/j.1365-2958.2007.05688.x. PMC 1891008. PMID 17462020.
- ^ a b c d e f g Kawano M (December 2012). "Divergently overlapping cis-encoded antisense RNA regulating toxin-antitoxin systems from E. coli: hok/sok, ldr/rdl, symE/symR" (PDF). RNA Biology. 9 (12): 1520–7. doi:10.4161/rna.22757. PMID 23131729.
- ^ Kawano M, Reynolds AA, Miranda-Rios J, Storz G (2005). "Detection of 5'- and 3'-UTR-derived small RNAs and cis-encoded antisense RNAs in Escherichia coli". Nucleic Acids Research. 33 (3): 1040–50. doi:10.1093/nar/gki256. PMC 549416. PMID 15718303.
- ^ a b c Fozo EM, Hemm MR, Storz G (December 2008). "Small toxic proteins and the antisense RNAs that repress them". Microbiology and Molecular Biology Reviews. 72 (4): 579–89, Table of Contents. doi:10.1128/MMBR.00025-08. PMC 2593563. PMID 19052321.
- ^ a b Brielle R, Pinel-Marie ML, Felden B (April 2016). "Linking bacterial type I toxins with their actions" (PDF). Current Opinion in Microbiology. Cell regulation. 30: 114–121. doi:10.1016/j.mib.2016.01.009. PMID 26874964.
- ^ Gerdes K, Wagner EG (April 2007). "RNA antitoxins". Current Opinion in Microbiology. 10 (2): 117–24. doi:10.1016/j.mib.2007.03.003. PMID 17376733.
- ^ The UniProt Consortium (2020). "UniProtKB - P39394 (SYME_ECOLI)". uniprot.org. Archived from the original on 10 July 2007. Retrieved 4 May 2020.
- ^ a b Waterhouse A, Bertoni M, Bienert S, Studer G, Tauriello G, Gumienny R, et al. (July 2018). "SWISS-MODEL: homology modelling of protein structures and complexes". Nucleic Acids Research. 46 (W1): W296–W303. doi:10.1093/nar/gky427. PMC 6030848. PMID 29788355.
- ^ a b Guex N, Peitsch MC, Schwede T (June 2009). "Automated comparative protein structure modeling with SWISS-MODEL and Swiss-PdbViewer: a historical perspective". Electrophoresis. 30 Suppl 1 (S1): S162-73. doi:10.1002/elps.200900140. PMID 19517507. S2CID 39507113.
- ^ a b Bienert S, Waterhouse A, de Beer TA, Tauriello G, Studer G, Bordoli L, Schwede T (January 2017). "The SWISS-MODEL Repository-new features and functionality". Nucleic Acids Research. 45 (D1): D313–D319. doi:10.1093/nar/gkw1132. PMC 5210589. PMID 27899672.
- ^ a b Studer G, Rempfer C, Waterhouse AM, Gumienny R, Haas J, Schwede T (April 2020). "QMEANDisCo-distance constraints applied on model quality estimation". Bioinformatics. 36 (8): 2647. doi:10.1093/bioinformatics/btaa058. PMC 7178391. PMID 32048708.
- ^ a b Bertoni M, Kiefer F, Biasini M, Bordoli L, Schwede T (September 2017). "Modeling protein quaternary structure of homo- and hetero-oligomers beyond binary interactions by homology". Scientific Reports. 7 (1): 10480. Bibcode:2017NatSR...710480B. doi:10.1038/s41598-017-09654-8. PMC 5585393. PMID 28874689.
- ^ Brenner SX, Miller JH. Encyclopedia of genetics. San Diego. ISBN 0-12-227080-0. OCLC 48655705.
Further reading
edit- Buts L, Lah J, Dao-Thi MH, Wyns L, Loris R (December 2005). "Toxin-antitoxin modules as bacterial metabolic stress managers". Trends in Biochemical Sciences. 30 (12): 672–9. doi:10.1016/j.tibs.2005.10.004. PMID 16257530.
- Gerdes K, Christensen SK, Løbner-Olesen A (May 2005). "Prokaryotic toxin-antitoxin stress response loci". Nature Reviews. Microbiology. 3 (5): 371–82. doi:10.1038/nrmicro1147. PMID 15864262. S2CID 13417307.
- Lewis LK, Harlow GR, Gregg-Jolly LA, Mount DW (August 1994). "Identification of high affinity binding sites for LexA which define new DNA damage-inducible genes in Escherichia coli". Journal of Molecular Biology. 241 (4): 507–23. doi:10.1006/jmbi.1994.1528. PMID 8057377.
- Christensen SK, Pedersen K, Hansen FG, Gerdes K (September 2003). "Toxin-antitoxin loci as stress-response-elements: ChpAK/MazF and ChpBK cleave translated RNAs and are counteracted by tmRNA". Journal of Molecular Biology. 332 (4): 809–19. doi:10.1016/S0022-2836(03)00922-7. PMID 12972253.
- Engelberg-Kulka H, Glaser G (1999). "Addiction modules and programmed cell death and antideath in bacterial cultures". Annual Review of Microbiology. 53: 43–70. doi:10.1146/annurev.micro.53.1.43. PMID 10547685.
- Cherepanov PP, Wackernagel W (May 1995). "Gene disruption in Escherichia coli: TcR and KmR cassettes with the option of Flp-catalyzed excision of the antibiotic-resistance determinant". Gene. 158 (1): 9–14. doi:10.1016/0378-1119(95)00193-A. PMID 7789817.
- Anantharaman V, Aravind L (2003). "New connections in the prokaryotic toxin-antitoxin network: relationship with the eukaryotic nonsense-mediated RNA decay system". Genome Biology. 4 (12): R81. doi:10.1186/gb-2003-4-12-r81. PMC 329420. PMID 14659018.