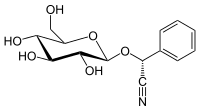
(R)-prunasin is a cyanogenic glycoside related to amygdalin. Chemically, it is the glucoside of (R)-mandelonitrile.
| |
| Names | |
|---|---|
| IUPAC name
(R)-(β-D-Glucopyranosyloxy)(phenyl)acetonitrile
| |
| Systematic IUPAC name
(R)-Phenyl{[(2R,3R,4S,5S,6R)-3,4,5-trihydroxy-6-(hydroxymethyl)oxan-2-yl]oxy}acetonitrile | |
| Other names
(R)-Prunasin
D-Prunasin D-Mandelonitrile-β-D-glucoside Prulaurasin Laurocerasin Sambunigrin | |
| Identifiers | |
3D model (JSmol)
|
|
| ChEBI | |
| ChemSpider | |
| ECHA InfoCard | 100.002.489 |
| EC Number |
|
| KEGG | |
PubChem CID
|
|
| UNII | |
CompTox Dashboard (EPA)
|
|
| |
| |
| Properties | |
| C14H17NO6 | |
| Molar mass | 295.291 g·mol−1 |
Except where otherwise noted, data are given for materials in their standard state (at 25 °C [77 °F], 100 kPa).
| |
Natural occurrences
editPrunasin is found in species in the genus Prunus such as Prunus japonica or P. maximowiczii and in bitter almonds.[1] It is also found in leaves and stems of Olinia ventosa, O. radiata, O. emarginata and O. rochetiana[2] and in Acacia greggii. It is a biosynthetic precursor of and intermediate in the biosynthesis of amygdalin, the chemical compound responsible for the taste of bitter almond.[citation needed]
It is also found in dandelion coffee, a coffee substitute.[citation needed]
Sambunigrin
editSambunigrin, a diastereomer of prunasin derived from (S)-mandelonitrile instead of it the (R)-isomer, has been isolated from leaves of the elder tree (Sambucus nigra).[3] Sambunigrin is present in the leaves and stems of elder at a 1:3 ratio of sambunigrin to prunasin, and 2:5 in the immature seed.[4] It is not found in the root.[4]
Biosynthesis
editOverview
edit(R)-prunasin begins with the common amino acid phenylalanine, which in plants is produced via the Shikimate pathway in primary metabolism. The pathway is catalyzed mainly by two cytochrome P450 (CYP) enzymes and a UDP-glucosyltransferase (UGT). After (R)-prunasin is formed, it is either converted into amygdalin by an additional UDP-glucosyltransferase or degraded into benzaldehyde and hydrogen cyanide.
Researchers have shown that the accumulation (or lack of) of prunasin and amygdalin in the almond kernel is responsible for sweet and bitter genotypes.[1] Because amygdalin is responsible for the bitter almond taste, almond growers have selected genotypes which minimize the biosynthesis of amygdalin. The CYP enzymes responsible for generation of prunasin are conserved across Prunus species.[5] There is a correlation between high concentration of prunasin in the vegetative regions of the plant and the sweetness of the almond, which is relevant to the almond agricultural industry. In almonds, the amygdalin biosynthetic genes are expressed at different levels in the tegument (mother tissue, or outer section) and cotyledon (kernel, or father tissue), and vary significantly during almond ontogeny.[1][6][7] The biosynthesis of prunasin occurs in the tegument, then transported to other tissues for conversion to amygdalin or degraded.[1][5]
Biosynthesis of (R)-prunasin
editBiosynthesis of (R)-prunasin in Prunus dulcis
editL-phenylalanine is first hydroxylated by CYP79D16, followed by a decarboxylation and dehydration, forming the E-oxime phenylacetaldoxime.[8] Next, CYP71AN24 catalyzes the rearrangement of the E-oxime to the Z-oxime followed by a dehydration and a hydroxylation to form mandelonitrile.[8] Finally, UGT85A19 or UGT94AF3 utilize UDP-glucose to glycosylate mandelonitrile, forming (R)-prunasin.[1]
After generating (R)-prunasin, the product is further glycosylated into amygdalin by either isoform UGT94AF1 or UGT94AF2.[1] Expression of UGTAF1/2 and prunasin hydrolases results in a low overall concentration of (R)-prunasin in almond tissues. It is important to note that an alpha-glucosidase or prunasin hydrolase can convert (R)-prunasin to mandelonitrile, its precursor, which can then be spontaneously or enzymatically hydrolyzed to benzaldehyde and hydrogen cyanide.[9]
Biosynthesis of (R)-prunasin in Eucalyptus cladocalyx
editThe biosynthesis of (R)-prunasin in E. cladocalyx, the sugar gum tree, has been shown to synthesize (R)-prunasin using an additional intermediate, phenylacetonitrile, using CYP706C55.[10] The pathway proceeds similarly to the pathway in Prunus species, where the multifunctional CYP79A125 catalyzes the conversion of L-phenylalanine to phenylacetaldoxime. Then, CYP706C55 catalyzes the dehydration of phenylacetaldoxime to phenylacetonitrile. Phenylacetonitrile is then hydroxylated by CYP71B103 to mandelonitrile. After generating mandelonitrile, UGT85A59 transfers glucose to yield (R)-prunasin.[10]
Metabolic Pathway Interactions
editAs (R)-prunasin is a product of secondary metabolism, its generation and degradation affect multiple metabolic pathways by consuming L-phenylalanine or increasing quantities of benzaldehyde and toxic hydrogen cyanide through prunasin degradation.
Metabolic profiling in almond, cassava, and sorghum identified a potential recycling mechanism where (R)-prunasin and other cyanogen glycosides may be utilized for nitrogen storage and nitrogen recycling without generating HCN.[11] In 2017, researchers used stable isotope labeling to demonstrate that 13C-labeled L-phenylalanine incorporated in (R)-prunasin could be converted to benzaldehyde and to salicylic acid using mandelonitrile as an intermediate.[12]
Toxicity
editThe toxicity of prunasin is based in its degradation products: (R)-prunasin is hydrolyzed to form benzaldehyde and hydrogen cyanide, which causes toxicity. Plants containing prunasin may therefore be toxic to animals, particularly ruminants.[13]
To degrade amygdalin to prunasin, amygdalin beta-glucosidase hydrolyzes the disaccharide to produce (R)-prunasin and D-glucose. Then, prunasin beta-glucosidase uses (R)-prunasin and water to produce D-glucose and mandelonitrile. After generating the aglycone mandelonitrile, then a mandelonitrile lyase can degrade the compound into benzaldehyde and hydrogen cyanide.[citation needed]
References
edit- ^ a b c d e f Sánchez-Pérez, Raquel; Belmonte, Fara Sáez; Borch, Jonas; Dicenta, Federico; Møller, Birger Lindberg; Jørgensen, Kirsten (April 2012). "Prunasin Hydrolases during Fruit Development in Sweet and Bitter Almonds". Plant Physiology. 158 (4): 1916–1932. doi:10.1104/pp.111.192021. ISSN 0032-0889. PMC 3320195. PMID 22353576.
- ^ Nahrstedt, Adolf; Rockenbach, Jürgen (1993). "Occurrence of the cyanogenic glucoside prunasin and II corresponding mandelic acid amide glucoside in Olinia species (oliniaceae)". Phytochemistry. 34 (2): 433. Bibcode:1993PChem..34..433N. doi:10.1016/0031-9422(93)80024-M.
- ^ Andrew Pengelly (2004), The Constituents of Medicinal Plants (2nd ed.), Allen & Unwin, pp. 44–45, ISBN 978-1-74114-052-1
- ^ a b Miller, Rebecca E.; Gleadow, Roslyn M.; Woodrow, Ian E. (2004). "Cyanogenesis in tropical Prunus turneriana: characterisation, variation and response to low light". Functional Plant Biology. 31 (5): 491–503. doi:10.1071/FP03218. ISSN 1445-4408. PMID 32688921.
- ^ a b Thodberg, Sara; Del Cueto, Jorge; Mazzeo, Rosa; Pavan, Stefano; Lotti, Concetta; Dicenta, Federico; Jakobsen Neilson, Elizabeth H.; Møller, Birger Lindberg; Sánchez-Pérez, Raquel (November 2018). "Elucidation of the Amygdalin Pathway Reveals the Metabolic Basis of Bitter and Sweet Almonds (Prunus dulcis)1[OPEN]". Plant Physiology. 178 (3): 1096–1111. doi:10.1104/pp.18.00922. ISSN 0032-0889. PMC 6236625. PMID 30297455.
- ^ Sánchez-Pérez, Raquel; Jørgensen, Kirsten; Olsen, Carl Erik; Dicenta, Federico; Møller, Birger Lindberg (March 2008). "Bitterness in Almonds". Plant Physiology. 146 (3): 1040–1052. doi:10.1104/pp.107.112979. ISSN 0032-0889. PMC 2259050. PMID 18192442.
- ^ Neilson, Elizabeth H.; Goodger, Jason Q.D.; Motawia, Mohammed Saddik; Bjarnholt, Nanna; Frisch, Tina; Olsen, Carl Erik; Møller, Birger Lindberg; Woodrow, Ian E. (December 2011). "Phenylalanine derived cyanogenic diglucosides from Eucalyptus camphora and their abundances in relation to ontogeny and tissue type". Phytochemistry. 72 (18): 2325–2334. Bibcode:2011PChem..72.2325N. doi:10.1016/j.phytochem.2011.08.022. PMID 21945721.
- ^ a b Yamaguchi, Takuya; Yamamoto, Kazunori; Asano, Yasuhisa (September 2014). "Identification and characterization of CYP79D16 and CYP71AN24 catalyzing the first and second steps in l-phenylalanine-derived cyanogenic glycoside biosynthesis in the Japanese apricot, Prunus mume Sieb. et Zucc". Plant Molecular Biology. 86 (1–2): 215–223. doi:10.1007/s11103-014-0225-6. ISSN 0167-4412. PMID 25015725. S2CID 14884838.
- ^ Zhou, Jiming; Hartmann, Stefanie; Shepherd, Brianne K.; Poulton, Jonathan E. (2002-07-01). "Investigation of the Microheterogeneity and Aglycone Specificity-Conferring Residues of Black Cherry Prunasin Hydrolases". Plant Physiology. 129 (3): 1252–1264. doi:10.1104/pp.010863. ISSN 0032-0889. PMC 166519. PMID 12114579.
- ^ a b Hansen, Cecilie Cetti; Sørensen, Mette; Veiga, Thiago A.M.; Zibrandtsen, Juliane F.S.; Heskes, Allison M.; Olsen, Carl Erik; Boughton, Berin A.; Møller, Birger Lindberg; Neilson, Elizabeth H.J. (November 2018). "Reconfigured Cyanogenic Glucoside Biosynthesis in Eucalyptus cladocalyx Involves a Cytochrome P450 CYP706C55". Plant Physiology. 178 (3): 1081–1095. doi:10.1104/pp.18.00998. ISSN 0032-0889. PMC 6236593. PMID 30297456.
- ^ Pičmanová, Martina; Neilson, Elizabeth H.; Motawia, Mohammed S.; Olsen, Carl Erik; Agerbirk, Niels; Gray, Christopher J.; Flitsch, Sabine; Meier, Sebastian; Silvestro, Daniele; Jørgensen, Kirsten; Sánchez-Pérez, Raquel (2015-08-01). "A recycling pathway for cyanogenic glycosides evidenced by the comparative metabolic profiling in three cyanogenic plant species". Biochemical Journal. 469 (3): 375–389. doi:10.1042/BJ20150390. ISSN 0264-6021. PMID 26205491. S2CID 206152311.
- ^ Diaz-Vivancos, Pedro; Bernal-Vicente, Agustina; Cantabella, Daniel; Petri, Cesar; Hern�ndez, Jos� Antonio (2017-12-01). "Metabolomics and Biochemical Approaches Link Salicylic Acid Biosynthesis to Cyanogenesis in Peach Plants". Plant and Cell Physiology. 58 (12): 2057–2066. doi:10.1093/pcp/pcx135. hdl:10317/7678. ISSN 0032-0781. PMID 29036663.
- ^ Peter R. Cheeke (1989). Toxicants of Plant Origin: Glycosides. Vol. 2. CRC Press. p. 137.