A coactivator is a type of transcriptional coregulator that binds to an activator (a transcription factor) to increase the rate of transcription of a gene or set of genes.[1] The activator contains a DNA binding domain that binds either to a DNA promoter site or a specific DNA regulatory sequence called an enhancer.[2][3] Binding of the activator-coactivator complex increases the speed of transcription by recruiting general transcription machinery to the promoter, therefore increasing gene expression.[3][4][5] The use of activators and coactivators allows for highly specific expression of certain genes depending on cell type and developmental stage.[2]
Some coactivators also have histone acetyltransferase (HAT) activity. HATs form large multiprotein complexes that weaken the association of histones to DNA by acetylating the N-terminal histone tail. This provides more space for the transcription machinery to bind to the promoter, therefore increasing gene expression.[1][4]
Activators are found in all living organisms, but coactivator proteins are typically only found in eukaryotes because they are more complex and require a more intricate mechanism for gene regulation.[1][4] In eukaryotes, coactivators are usually proteins that are localized in the nucleus.[1][6]
Mechanism
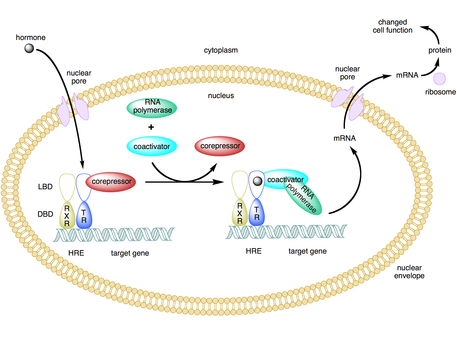
editSome coactivators indirectly regulate gene expression by binding to an activator and inducing a conformational change that then allows the activator to bind to the DNA enhancer or promoter sequence.[2][7][8] Once the activator-coactivator complex binds to the enhancer, RNA polymerase II and other general transcription machinery are recruited to the DNA and transcription begins.[9]
Histone acetyltransferase
editNuclear DNA is normally wrapped tightly around histones, making it hard or impossible for the transcription machinery to access the DNA. This association is due primarily to the electrostatic attraction between the DNA and histones as the DNA phosphate backbone is negatively charged and histones are rich in lysine residues, which are positively charged.[10] The tight DNA-histone association prevents the transcription of DNA into RNA.
Many coactivators have histone acetyltransferase (HAT) activity meaning that they can acetylate specific lysine residues on the N-terminal tails of histones.[4][7][11] In this method, an activator binds to an enhancer site and recruits a HAT complex that then acetylates nucleosomal promoter-bound histones by neutralizing the positively charged lysine residues.[7][11] This charge neutralization causes the histones to have a weaker bond to the negatively charged DNA, which relaxes the chromatin structure, allowing other transcription factors or transcription machinery to bind to the promoter (transcription initiation).[4][11] Acetylation by HAT complexes may also help keep chromatin open throughout the process of elongation, increasing the speed of transcription.[4]
Acetylation of the N-terminal histone tail is one of the most common protein modifications found in eukaryotes, with about 85% of all human proteins being acetylated.[12] Acetylation is crucial for synthesis, stability, function, regulation and localization of proteins and RNA transcripts.[11][12]
HATs function similarly to N-terminal acetyltransferases (NATs) but their acetylation is reversible unlike in NATs.[13] HAT mediated histone acetylation is reversed using histone deacetylase (HDAC), which catalyzes the hydrolysis of lysine residues, removing the acetyl group from the histones.[4][7][11] This causes the chromatin to close back up from their relaxed state, making it difficult for the transcription machinery to bind to the promoter, thus repressing gene expression.[4][7]
Examples of coactivators that display HAT activity include CARM1, CBP and EP300.[14][15]
Corepression
editMany coactivators also function as corepressors under certain circumstances.[5][9] Cofactors such as TAF1 and BTAF1 can initiate transcription in the presence of an activator (act as a coactivator) and repress basal transcription in the absence of an activator (act as a corepressor).[9]
Significance
editBiological significance
editTranscriptional regulation is one of the most common ways for an organism to alter gene expression.[16] The use of activation and coactivation allows for greater control over when, where and how much of a protein is produced.[1][7][16] This enables each cell to be able to quickly respond to environmental or physiological changes and helps to mitigate any damage that may occur if it were otherwise unregulated.[1][7]
Associated disorders
editMutations to coactivator genes leading to loss or gain of protein function have been linked to diseases and disorders such as birth defects, cancer (especially hormone dependent cancers), neurodevelopmental disorders and intellectual disability (ID), among many others.[17][5] Dysregulation leading to the over- or under-expression of coactivators can detrimentally interact with many drugs (especially anti-hormone drugs) and has been implicated in cancer, fertility issues and neurodevelopmental and neuropsychiatric disorders.[5] For a specific example, dysregulation of CREB-binding protein (CBP)—which acts as a coactivator for numerous transcription factors within the central nervous system (CNS), reproductive system, thymus and kidneys—has been linked to Huntington's disease, leukaemia, Rubinstein-Taybi syndrome, neurodevelopmental disorders and deficits of the immune system, hematopoiesis and skeletal muscle function.[14][18]
As drug targets
editCoactivators are promising targets for drug therapies in the treatment of cancer, metabolic disorder, cardiovascular disease and type 2 diabetes, along with many other disorders.[5][19] For example, the steroid receptor coactivator (SCR) NCOA3 is often overexpressed in breast cancer, so the development of an inhibitor molecule that targets this coactivator and decreases its expression could be used as a potential treatment for breast cancer.[15][20]
Because transcription factors control many different biological processes, they are ideal targets for drug therapy.[14][21] The coactivators that regulate them can be easily replaced with a synthetic ligand that allows for control over an increase or decrease in gene expression.[14]
Further technological advances will provide new insights into the function and regulation of coactivators at a whole-organism level and elucidate their role in human disease, which will hopefully provide better targets for future drug therapies.[14][15]
Known coactivators
editTo date there are more than 300 known coregulators.[15] Some examples of these coactivators include:[22]
- ARA54 targets androgen receptors
- ATXN7L3 targets several members of the nuclear receptor superfamily
- BCL3 targets 9-cis retinoic acid receptor (RXR)
- CBP targets many transcription factors
- CDC25B targets steroid receptors
- COPS5 targets several nuclear receptors
- DDC targets androgen receptors
- EP300 targets many transcription factors
- KAT5 targets many nuclear receptors
- KDM1A targets androgen receptors
- Steroid receptor coactivator (SRC) family
- YAP targets transcription factors
- WWTR1 targets transcription factors
See also
editReferences
edit- ^ a b c d e f Courey AJ (2008). Mechanisms in transcriptional regulation. Malden, MA: Blackwell. ISBN 978-1-4051-0370-1. OCLC 173367793.
- ^ a b c "General transcription factor / transcription factor". Learn Science at Scitable. Retrieved 2017-11-29.
- ^ a b Pennacchio LA, Bickmore W, Dean A, Nobrega MA, Bejerano G (April 2013). "Enhancers: five essential questions". Nature Reviews Genetics. 14 (4): 288–95. doi:10.1038/nrg3458. PMC 4445073. PMID 23503198.
- ^ a b c d e f g h Brown CE, Lechner T, Howe L, Workman JL (January 2000). "The many HATs of transcription coactivators". Trends in Biochemical Sciences. 25 (1): 15–9. doi:10.1016/S0968-0004(99)01516-9. PMID 10637607.
- ^ a b c d e Kumar R, O'Malley BW (2008). NR coregulators and human diseases. Hackensack, N.J.: World Scientific. ISBN 978-981-270-536-5. OCLC 261137374.
- ^ Vosnakis N, Koch M, Scheer E, Kessler P, Mély Y, Didier P, Tora L (September 2017). "Coactivators and general transcription factors have two distinct dynamic populations dependent on transcription". The EMBO Journal. 36 (18): 2710–2725. doi:10.15252/embj.201696035. PMC 5599802. PMID 28724529.
- ^ a b c d e f g Spiegelman BM, Heinrich R (October 2004). "Biological control through regulated transcriptional coactivators". Cell. 119 (2): 157–67. doi:10.1016/j.cell.2004.09.037. PMID 15479634. S2CID 14668705.
- ^ Scholes NS, Weinzierl RO (May 2016). "Molecular Dynamics of "Fuzzy" Transcriptional Activator-Coactivator Interactions". PLOS Computational Biology. 12 (5): e1004935. Bibcode:2016PLSCB..12E4935S. doi:10.1371/journal.pcbi.1004935. PMC 4866707. PMID 27175900.
- ^ a b c Thomas MC, Chiang CM (May 2006). "The general transcription machinery and general cofactors". Critical Reviews in Biochemistry and Molecular Biology. 41 (3): 105–78. CiteSeerX 10.1.1.376.5724. doi:10.1080/10409230600648736. PMID 16858867. S2CID 13073440.
- ^ Decher, Gero (1997-08-29). "Fuzzy Nanoassemblies: Toward Layered Polymeric Multicomposites". Science. 277 (5330): 1232–1237. doi:10.1126/science.277.5330.1232. ISSN 0036-8075.
- ^ a b c d e Hermanson O, Glass CK, Rosenfeld MG (2002). "Nuclear receptor coregulators: multiple modes of modification". Trends in Endocrinology and Metabolism. 13 (2): 55–60. doi:10.1016/s1043-2760(01)00527-6. PMID 11854019. S2CID 38649132.
- ^ a b Van Damme P, Hole K, Pimenta-Marques A, Helsens K, Vandekerckhove J, Martinho RG, Gevaert K, Arnesen T (July 2011). "NatF contributes to an evolutionary shift in protein N-terminal acetylation and is important for normal chromosome segregation". PLOS Genetics. 7 (7): e1002169. doi:10.1371/journal.pgen.1002169. PMC 3131286. PMID 21750686.
- ^ Starheim KK, Gevaert K, Arnesen T (April 2012). "Protein N-terminal acetyltransferases: when the start matters". Trends in Biochemical Sciences. 37 (4): 152–61. doi:10.1016/j.tibs.2012.02.003. PMID 22405572.
- ^ a b c d e Lonard DM, O'Malley BW (October 2012). "Nuclear receptor coregulators: modulators of pathology and therapeutic targets". Nature Reviews. Endocrinology. 8 (10): 598–604. doi:10.1038/nrendo.2012.100. PMC 3564250. PMID 22733267.
- ^ a b c d Hsia EY, Goodson ML, Zou JX, Privalsky ML, Chen HW (October 2010). "Nuclear receptor coregulators as a new paradigm for therapeutic targeting". Advanced Drug Delivery Reviews. 62 (13): 1227–37. doi:10.1016/j.addr.2010.09.016. PMC 5004779. PMID 20933027.
- ^ a b "Enhancer". Learn Science at Scitable. Retrieved 2017-11-29.
- ^ Berk A, Zipursky SL, Matsudaira PT, Baltimore D, Darnell J (2000). Lodish HF (ed.). Molecular Cell Biology (4th ed.). New York: W.H. Freeman. ISBN 978-0-7167-3136-8. OCLC 41266312.
- ^ Becnel LB, Darlington YF, Orechsner S, Easton-Marks J, Watkins CA, McOwiti A, Kankanamge WH, Dehart M, Silva CM. "CBP". NURSA Molecules. doi:10.1621/8egsudafco.
- ^ "Nuclear Receptors". courses.washington.edu. Retrieved 2017-11-29.
- ^ Tien JC, Xu J (November 2012). "Steroid receptor coactivator-3 as a potential molecular target for cancer therapy". Expert Opinion on Therapeutic Targets. 16 (11): 1085–96. doi:10.1517/14728222.2012.718330. PMC 3640986. PMID 22924430.
- ^ Sladek FM (October 2003). "Nuclear receptors as drug targets: new developments in coregulators, orphan receptors and major therapeutic areas". Expert Opinion on Therapeutic Targets. 7 (5): 679–84. doi:10.1517/14728222.7.5.679. PMID 14498828. S2CID 16891519.
- ^ "NURSA - Molecules". nursa.org. Retrieved 2017-11-30.
External links
edit- Nuclear Receptor Signalling Atlas (NIH-funded research consortium and database; includes open-access PubMed-indexed journal, Nuclear Receptor Signaling)
- TcoF - Dragon database of transcription co-factors and transcription factor interacting proteins