Illegitimate recombination, or nonhomologous recombination, is the process by which two unrelated double stranded segments of DNA are joined. This insertion of genetic material which is not meant to be adjacent tends to lead to genes being broken causing the protein which they encode to not be properly expressed. One of the primary pathways by which this will occur is the repair mechanism known as non-homologous end joining (NHEJ).[1]
Discovery
editIllegitimate recombination is a natural process which was first found to be present within E. coli. A 700-1400 base pair segment of DNA was found to have inserted itself into the gal and lac operons resulting in a strong polar mutation.[2] This mechanism was then found to have the ability to insert other short genetic sequences into other locations within the bacterial genome often leading to a change in the expression of neighboring genes. Oftentimes it leads to the neighboring genes to simply shut off. However some of these segments also had strong start and stop signals which changed the regulation of neighboring genes leading in changes in the amount of transcription. What differentiated this form of genetic recombination from those dependent of genetic homology was that the process observed as illegitimate did not require the use of homologous segments of DNA. While not being entirely understood at the time, it was recognized to hold potential in generating changes in the chromosomal evolution.[3]
Mechanism
editIn prokaryotes
editIn prokaryotes, illegitimate recombination results in a mutation of the genetic sequence of the prokaryote. This process takes different forms in eukaryotes one of which is deletions. In a deletion mutation the prokaryotic organism undergoes illegitimate recombination resulting in the removal of a continuous segment of genetic code. However this form of mutation occurs infrequently among mutants of natural origin rather than those that have been induced. Another form of illegitimate recombination in prokaryotes is that of a duplication mutations of a genome. In this case a portion of the parental genome is inserted multiple times into the genome. This duplication either inserts the genetic material in the same orientation or opposite of the original parental segments as it is non-homology driven.[3]
In eukaryotes
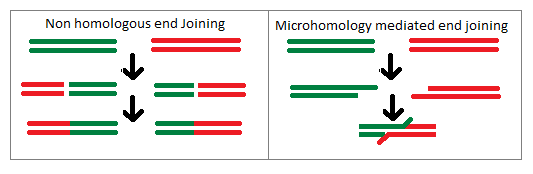
editThe mechanism of Illegitimate recombination is that of non-homologous end joining in which two strands of DNA not sharing homology will be joined together by the gene repair machinery. Upon recognition of a double strand break a protein complex will keep the two strands within close enough proximity in order to allow for repair of the strands. Next the ends of the DNA are repaired such that any incorrect or damaged nucleotides are removed. Once this happens the strands are able to be ligated together such that they are a single strand of DNA which previously had not been adjacent. This process is common for eukaryotic cells and tends to act as a repair mechanism, but can lead to these mutations if illegitimate recombination occurs. The illegitimate recombination will often take the form of large chromosomal aberrations within a eukaryotic organism as it has much larger segments of DNA than prokaryotic cells. As of such non-homologous end joining can cause illegitimate recombination which creates insertion and deletion mutations in chromosomes as well as translocation of one chromosomal segment to that of another chromosome. These large scale changes in the chromosome in eukaryotic organisms tend to have deleterious effects on the organism rather than conferring a type of genetic advantage.[4]
Deleterious effects on organisms
editIllegitimate recombination oftentimes has deleterious effects on an organism as it results in a large scale change on the genetic sequence of an organism. These changes will result in mutations as the joining of DNA not based on homology will most often place genetic elements in locations in which they previously had not been placed. This can disrupt the function of genes which may be essential to the function of an organism. In the case of cancer it has been found that tumors can be a result of illegitimate recombination resulting in hairpin formation which alters the gene function within the genome of tumor cells.[5]
Applications
editIllegitimate recombination is a tool which can be used in the laboratory as well as it is a useful research tool. Illegitimate recombination can generate random mutagenesis in order to generate a random alteration of the genetic sequence of an organism.[6] The induction of this mutagenesis allows for the study of a genetic sequence by creating a mutation in a genetic segment altering the function of that genetic segment. This allows for the study of gene function through the analysis of differences between mutants and natural organisms to interpret what process a gene is linked to.
References
edit- ^ Wilson TE (2006). "Nonhomologous end-joining: Mechanisms, conservation and relationship to illegitimate recombination". In Aguilera A, Rothstein R (eds.). Molecular Genetics of Recombination. Topics in Current Genetics. Vol. 17. Berlin, Heidelberg: Springer. pp. 487–513. doi:10.1007/978-3-540-71021-9_17. ISBN 978-3-540-71020-2.
- ^ Sherratt D (July 1978). "Illegitimate recombination legitimised". Nature. 274 (5668): 213–4. Bibcode:1978Natur.274..213S. doi:10.1038/274213a0. PMID 355886.
- ^ a b Weisberg RA, Adhya S (December 1977). "Illegitimate recombination in bacteria and bacteriophage". Annual Review of Genetics. 11 (1): 451–73. doi:10.1146/annurev.ge.11.120177.002315. PMID 339822.
- ^ Ehrlich SD, Bierne H, d'Alençon E, Vilette D, Petranovic M, Noirot P, Michel B (December 1993). "Mechanisms of illegitimate recombination". Gene. 135 (1–2): 161–6. doi:10.1016/0378-1119(93)90061-7. PMID 8276254.
- ^ Zucman-Rossi J, Legoix P, Victor JM, Lopez B, Thomas G (September 1998). "Chromosome translocation based on illegitimate recombination in human tumors". Proceedings of the National Academy of Sciences of the United States of America. 95 (20): 11786–91. Bibcode:1998PNAS...9511786Z. doi:10.1073/pnas.95.20.11786. PMC 21718. PMID 9751743.
- ^ Khattak FA, Kumar A, Kamal E, Kunisch R, Lewin A (September 2012). "Illegitimate recombination: an efficient method for random mutagenesis in Mycobacterium avium subsp. hominissuis". BMC Microbiology. 12: 204. doi:10.1186/1471-2180-12-204. PMC 3511198. PMID 22966811.