The two-domain system is a biological classification by which all organisms in the tree of life are classified into two domains, Bacteria and Archaea.[1][2][3] It emerged from development of knowledge of archaea diversity and challenges to the widely accepted three-domain system that classifies life into Bacteria, Archaea, and Eukarya.[4] It was preceded by the eocyte hypothesis of James A. Lake in the 1980s,[5] which was largely superseded by the three-domain system, due to evidence at the time.[6] Better understanding of archaea, especially of their roles in the origin of eukaryotes through symbiogenesis with bacteria, led to the revival of the eocyte hypothesis in the 2000s.[7][8] The two-domain system became more widely accepted after the discovery of a large group (superphylum) of archaea called Asgard in 2017,[9] which evidence suggests to be the evolutionary root of eukaryotes, thereby making eukaryotes members of the domain Archaea.[10]
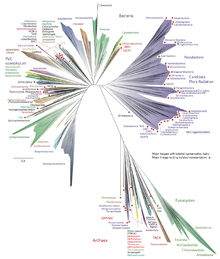
While the features of Asgard archaea do not completely rule out the three-domain system,[11][12] the notion that eukaryotes originated within Archaea has been strengthened by genetic and proteomic studies.[13] Under the three-domain system, Eukarya is mainly distinguished by the presence of "eukaryotic signature proteins" that are not found in Archaea and Bacteria. However, Asgard archaea contain genes that code for multiple such proteins.[3]
Background edit
Classification of life into two main divisions is not a new concept, with the first such proposal by French biologist Édouard Chatton in 1938. Chatton distinguished organisms into:
- Procaryotes (including bacteria)
- Eucaryotes (including protozoans)[14]
These were later named empires, and Chatton's classification as the two-empire system.[15] Chatton used the name Eucaryotes only for protozoans, excluded other eukaryotes, and published in limited circulation so that his work was not recognised. His classification was rediscovered by Canadian bacteriologist Roger Yates Stanier of the University of California in Berkeley in 1961 while at the Pasteur Institute in Paris.[14] The next year, Stanier and his colleague Cornelis Bernardus van Niel published in Archiv für Mikrobiologie (now Archives of Microbiology) Chatton's classification with Eucaryotes eloborated to include higher algae, protozoans, fungi, plants and animals.[16] It became a popular system of classification, as John O. Corliss wrote in 1986: "[The] Chatton-Stanier concept of a kingdom (better, superkingdom) Prokaryota for bacteria (in the broadest sense) and a second superkingdom Eukaryota for all other organisms has been widely accepted with enthusiasm."[17]
In 1977, Carl Woese and George E. Fox classified prokaryotes into two groups (kingdoms), Archaebacteria (for methanogens, the first known archaea) and Eubacteria, based on their 16S ribosomal RNA (16S rRNA) genes.[18] In 1984, James A. Lake, Michael W. Clark, Eric Henderson, and Melanie Oakes of the University of California, Los Angeles described what was known as "a group of sulfur-dependent bacteria" as a new group of organisms called eocytes (for "dawn cells") and created a new kingdom Eocyta. With it they proposed the existence of four kingdoms, based on the structure and composition of the ribosomal subunits, namely Archaebacteria, Eubacteria, Eukaryote and Eocyta [19] Lake further analysed the rRNA sequences of the four groups and suggested that eukaryotes originated from eocytes, and not archaebacteria, as was generally assumed.[20] This was the basis of the eocyte hypothesis.[6] In 1988, he proposed the division of all life forms into two taxonomic groups:[5]
- Karyotes (that include eukaryotes and proto-eukaryotic organisms such as eocytes)
- Parkaryotes (that consist of eubacteria and archaea such as halobacteria and methanogens[21]
In 1990, Woese, Otto Kandler and Mark Wheelis showed that archaea are distinct group of organisms and that eocytes (renamed Crenarchaeota as a phylum of Archaea[22] but corrected as Thermoproteota in 2021[23]) are Archaea. They introduced the major division of life into the three-domain system comprising domain Eucarya, domain Bacteria and domain Archaea.[24] With a number of revisions of details and discoveries of several new archaea lineages, Woese's classification gradually gained acceptance as "arguably the best-developed and most widely-accepted scientific hypotheses [with the five-kingdom classification] regarding the evolutionary history of life."[25]
The three-domain concept did not, however, resolve the issues with the relationship between archaea and eukaryotes.[12][26] As Ford Doolittle, then at the Dalhousie University, put it in 2020: "[The] three-domain tree wrongly represents evolutionary relationships, presenting a misleading view about how eukaryotes evolved from prokaryotes. The three-domain tree does recognize a specific archaeal–eukaryotic affinity, but it would have the latter arising independently, not from within, the former."[4]
Concept edit
The two-domain system relies mainly on two key concepts that define eukaryotes as members of the domain Archaea and not as a separate domain: eukaryotes originated within Archaea, and Asgards represent the origin of eukaryotes.[27][28]
Eukaryotes evolved from archaea edit
The three-domain system presumes that eukaryotes are more closely related to archaea than to bacteria and are sister group to archaea, thus, it treats them as separate domain.[29] As more new archaea were discovered in the early 2000s, this distinction became doubtful as eukaryotes became deeply nested within archaea. The origin of eukaryotes from archaea, meaning the two are of the same group, came to be supported by studies based on ribosome protein sequencing and phylogenetic analyses in 2004.[30][31] Phylogenomic analysis of about 6000 gene sets from 185 bacterial, archaeal and eukaryotic genomes in 2007 also suggested origin of eukaryotes from Euryarchaeota (specifically the Thermoplasmatales).[32]
In 2008, researchers from Natural History Museum, London and Newcastle University reported a comprehensive analysis of 53 genes from archaea, bacteria and eukaryotes that included essential components of the nucleic acid replication, transcription, and translation machineries. The conclusion was that eukaryotes evolved from archaea, specifically Crenarchaeota (eocytes) and the results "favor a topology that supports the eocyte hypothesis rather than archaebacterial monophyly and the 3-domains tree of life."[26] A study around the same time also found several genes common to eukaryotes and Crenarchaeota.[33] These accumulating evidences support the two-domain system.[22]
One of the distinctions of the domain Eukarya in the three-domain system is that eukaryotes have unique proteins such as actin (cytoskeletal microfilament involved in cell motility), tubulin (component of the large cytoskeleton, microtubule) and the ubiquitin system (protein degradation and recycling) that are not found in prokaryotes. However, these so-called "eukaryotic signature proteins"[3] are encoded in genomes of TACK (comprising the phyla Thaumarchaeota, Aigarchaeota, Crenarchaeota and Korarchaeota) archaea, but not encoded in other archaea genomes.[22] The first eukaryotic proteins identified in Crenarchaeota were actin and actin-related proteins (Arp) 2 and 3, perhaps explaining the origin of eukaryotes by symbiogenic phagocytosis, in which an ancient archaeal host had an actin based mechanism by which to envelop other cells, like protomitochondrial bacteria.[34]
Tubulin-like proteins named artubulins are found in the genomes of several ammonium-oxidising Thaumarchaeota.[35] Endosomal sorting complexes, required for transport (ESCRT III), involved in eukaryotic cell division, are found in all TACK groups.[36] The ESCRT-III-like proteins constitute the primary cell division system in these archaea.[37][38] Genes encoding the ubiquitin system are known from multiple genomes of Aigarchaeota.[39] Ubiquitin-related protein called Urm1 is also present in Crenarchaeota.[40] DNA replication system (GINS proteins) in Crenarchaeota and Halobacteria are similar to the CMG (CDC45, MCM, GINS) complex of eukaryotes.[41] The presence of these eukaryotic proteins in archaea indicates their direct relationship and that eukaryotes emerged from archaea.[22][42]
Asgards are the last eukaryotic common ancestor edit
The discovery of Asgard, described as "eukaryote-like archaea",[43] in 2012[44][45] and the following phylogenetic analyses have strengthened the two-domain view of life.[46] Asgard Archaea called Lokiarchaeota contain even more eukaryotic protein-genes than the TACK group. Initial genetic analysis and later reanalysis showed that out of over 31 selected eukaryotic genes in the archaea, 75% of them directly support eukaryote-archaea grouping, meaning a single domain of Archaea including eukaryotes;[47][48] although the findings did not completely rule out the three-domain system.[49]
As more Asgard groups were subsequently discovered including Thorarchaeota, Odinarchaeota and Heimdallarchaeota, their relationships with eukaryotes became better established. Phylogenetic analyses using ribosomal RNA genes indicated that eukaryotes stemmed from Asgards, and that Heimdallarchaeota are the closest relatives of eukaryotes.[9][50] Eukaryotic origin from Heimdallarchaeota is also supported by phylogenomic study in 2020.[13] A new group of Asgard found in 2021 (provisionally named Wukongarchaeota) also indicated a deep root for eukaryotic origin.[51] A report in 2022 of another Asgard, named Njordarchaeota, indicates that Heimdallarchaeota-Wukongarchaeota branch is possibly the origin group for eukaryotes.[52]
The Asgards contain at least 80 genes for eukaryotic signature proteins.[53] In addition to actin, tubulin, ubiquitin and ESCRT proteins found in TACK archaea, Asgards contain functional genes for several other eukaryotic proteins such as profilins,[54] ubiquitin system (E1-like, E2-like and small-RING finger (srfp) proteins),[55] membrane-trafficking systems (such as Sec23/24 and TRAPP domains), a variety of small GTPases[46] (including Gtr/Rag family GTPase orthologues[56]), and gelsolins.[57] Although this information do not completely resolve the three-domain and two-domain controversies,[43] they are generally considered to favour the two-domain system.[3][13][58]
Classification edit
The two-domain system defines classification of all known cellular life forms into two domains: Bacteria and Archaea. It overrides the domain Eukaryota recognised in the three-domain classification as one of the main domains. In contrast to the eocyte hypothesis, which proposed two major groups of life (similar to domains) and posited that archaea could be divided to both bacterial and eukaryotic groups, it merged archaea and eukaryotes into a single domain, bacteria entirely in a separate domain.[4]
Domain Bacteria edit
It consists of all bacteria, which are prokaryotes (lacking nucleus), thus, Domain Bacteria is made up solely of prokaryotic organisms.[59][60] Some examples are:
- Cyanobacteria – photosynthesising bacteria related to the plastids of eukaryotes.[61]
- Spirochaetota – Gram-negative bacteria involved in human diseases like syphilis and lyme disease.[62]
- Actinomycetota – Gram-positive bacteria including Streptomyces species from which several antibiotics are derived including streptomycin, neomycin, bottromycins and chloramphenicol.[63][64]
Domain Archaea edit
It comprises both prokaryotic and eukaryotic organisms.[65]
Archaea
Archaea are prokaryotic organisms, some examples are:
- All methanogens – which produce the gas methane.
- Most halophiles – which live in very salty water.
- Most thermoacidophiles – which live in acidic high-temperature water.
Eukarya
Eukaryotes have a nucleus in their cells, and include:
References edit
- ^ Bolshoy, Alexander; Volkovich, Zeev (Vladimir); Kirzhner, Valery; Barzily, Zeev (2010), "Biological Classification", Genome Clustering, Studies in Computational Intelligence, vol. 286, Berlin, Heidelberg: Springer Berlin Heidelberg, pp. 17–22, doi:10.1007/978-3-642-12952-0_2, ISBN 978-3-642-12951-3, retrieved 2022-05-14
- ^ Raymann, Kasie; Brochier-Armanet, Céline; Gribaldo, Simonetta (2015). "The two-domain tree of life is linked to a new root for the Archaea". Proceedings of the National Academy of Sciences of the United States of America. 112 (21): 6670–6675. Bibcode:2015PNAS..112.6670R. doi:10.1073/pnas.1420858112. PMC 4450401. PMID 25964353.
- ^ a b c d Nobs, Stephanie-Jane; MacLeod, Fraser I.; Wong, Hon Lun; Burns, Brendan P. (2022). "Eukarya the chimera: eukaryotes, a secondary innovation of the two domains of life?". Trends in Microbiology. 30 (5): 421–431. doi:10.1016/j.tim.2021.11.003. PMID 34863611. S2CID 244823103.
- ^ a b c Doolittle, W. Ford (2020). "Evolution: Two Domains of Life or Three?". Current Biology. 30 (4): R177–R179. doi:10.1016/j.cub.2020.01.010. PMID 32097647.
- ^ a b Lake, James A. (1988). "Origin of the eukaryotic nucleus determined by rate-invariant analysis of rRNA sequences". Nature. 331 (6152): 184–186. Bibcode:1988Natur.331..184L. doi:10.1038/331184a0. PMID 3340165. S2CID 4368082.
- ^ a b Archibald, John M. (2008). "The eocyte hypothesis and the origin of eukaryotic cells". Proceedings of the National Academy of Sciences. 105 (51): 20049–20050. Bibcode:2008PNAS..10520049A. doi:10.1073/pnas.0811118106. PMC 2629348. PMID 19091952.
- ^ Poole, Anthony M.; Penny, David (2007). "Evaluating hypotheses for the origin of eukaryotes". BioEssays. 29 (1): 74–84. doi:10.1002/bies.20516. PMID 17187354.
- ^ Foster, Peter G.; Cox, Cymon J.; Embley, T. Martin (2009). "The primary divisions of life: a phylogenomic approach employing composition-heterogeneous methods". Philosophical Transactions of the Royal Society of London. Series B, Biological Sciences. 364 (1527): 2197–2207. doi:10.1098/rstb.2009.0034. PMC 2873002. PMID 19571240.
- ^ a b Zaremba-Niedzwiedzka, Katarzyna; Caceres, Eva F.; Saw, Jimmy H.; Bäckström, Disa; Juzokaite, Lina; Vancaester, Emmelien; Seitz, Kiley W.; Anantharaman, Karthik; Starnawski, Piotr (2017). "Asgard archaea illuminate the origin of eukaryotic cellular complexity" (PDF). Nature. 541 (7637): 353–358. Bibcode:2017Natur.541..353Z. doi:10.1038/nature21031. OSTI 1580084. PMID 28077874. S2CID 4458094.
- ^ Eme, Laura; Spang, Anja; Lombard, Jonathan; Stairs, Courtney W.; Ettema, Thijs J. G. (10 November 2017). "Archaea and the origin of eukaryotes". Nature Reviews Microbiology. 15 (12): 711–723. doi:10.1038/nrmicro.2017.133. ISSN 1740-1534. PMID 29123225. S2CID 8666687.
- ^ Da Cunha, Violette; Gaia, Morgan; Nasir, Arshan; Forterre, Patrick (2018). "Asgard archaea do not close the debate about the universal tree of life topology". PLOS Genetics. 14 (3): e1007215. doi:10.1371/journal.pgen.1007215. PMC 5875737. PMID 29596428.
- ^ a b Zhou, Zhichao; Liu, Yang; Li, Meng; Gu, Ji-Dong (2018). "Two or three domains: a new view of tree of life in the genomics era". Applied Microbiology and Biotechnology. 102 (7): 3049–3058. doi:10.1007/s00253-018-8831-x. PMID 29484479. S2CID 3541409.
- ^ a b c Williams, Tom A.; Cox, Cymon J.; Foster, Peter G.; Szöllősi, Gergely J.; Embley, T. Martin (2020). "Phylogenomics provides robust support for a two-domains tree of life". Nature Ecology & Evolution. 4 (1): 138–147. doi:10.1038/s41559-019-1040-x. PMC 6942926. PMID 31819234.
- ^ a b Katscher, Friedrich (2004). "The History of the Terms Prokaryotes and Eukaryotes". Protist. 155 (2): 257–263. doi:10.1078/143446104774199637. PMID 15305800.
- ^ Mayr, Ernst (1998). "Two empires or three?". Proceedings of the National Academy of Sciences. 95 (17): 9720–9723. Bibcode:1998PNAS...95.9720M. doi:10.1073/pnas.95.17.9720. PMC 33883. PMID 9707542.
- ^ Stanier, R. Y.; Van Niel, C. B. (1962). "The concept of a bacterium" (PDF). Archiv für Mikrobiologie. 42: 17–35. doi:10.1007/BF00425185. PMID 13916221. S2CID 29859498.
- ^ Corliss, John O. (1986). "The Kingdoms of Organisms: From a Microscopist's Point of View". Transactions of the American Microscopical Society. 105 (1): 1–10. doi:10.2307/3226544. JSTOR 3226544.
- ^ Woese CR, Fox GE (November 1977). "Phylogenetic structure of the prokaryotic domain: the primary kingdoms". Proceedings of the National Academy of Sciences of the United States of America. 74 (11): 5088–90. Bibcode:1977PNAS...74.5088W. doi:10.1073/pnas.74.11.5088. PMC 432104. PMID 270744.
- ^ Lake, James A.; Henderson, Eric; Oakes, Melanie; Clark, Michael W. (June 1984). "Eocytes: A new ribosome structure indicates a kingdom with a close relationship to eukaryotes". PNAS. 81 (12): 3786–3790. Bibcode:1984PNAS...81.3786L. doi:10.1073/pnas.81.12.3786. PMC 345305. PMID 6587394.
- ^ Lake, J.A. (1987). "Prokaryotes and Archaebacteria Are Not Monophyletic: Rate Invariant Analysis of rRNA Genes Indicates That Eukaryotes and Eocytes Form a Monophyletic Taxon". Cold Spring Harbor Symposia on Quantitative Biology. 52: 839–846. doi:10.1101/SQB.1987.052.01.091. ISSN 0091-7451. PMID 3454292.
- ^ Lake, James A. (1991). "Tracing origins with molecular sequences: metazoan and eukaryotic beginnings". Trends in Biochemical Sciences. 16 (2): 46–50. doi:10.1016/0968-0004(91)90020-V. PMID 1858129.
- ^ a b c d Koonin, Eugene V. (2015). "Origin of eukaryotes from within archaea, archaeal eukaryome and bursts of gene gain: eukaryogenesis just made easier?". Philosophical Transactions of the Royal Society of London. Series B, Biological Sciences. 370 (1678): 20140333. doi:10.1098/rstb.2014.0333. PMC 4571572. PMID 26323764.
- ^ Oren, Aharon; Garrity, George M. (2021). "Valid publication of the names of forty-two phyla of prokaryotes". International Journal of Systematic and Evolutionary Microbiology. 71 (10): Online. doi:10.1099/ijsem.0.005056. ISSN 1466-5034. PMID 34694987. S2CID 239887308.
- ^ Woese CR, Kandler O, Wheelis ML (June 1990). "Towards a natural system of organisms: proposal for the domains Archaea, Bacteria, and Eucarya". Proceedings of the National Academy of Sciences of the United States of America. 87 (12): 4576–9. Bibcode:1990PNAS...87.4576W. doi:10.1073/pnas.87.12.4576. PMC 54159. PMID 2112744.
- ^ Case, Emily (2008). "Teaching Taxonomy: How Many Kingdoms?". The American Biology Teacher. 70 (8): 472–477. doi:10.1662/0002-7685(2008)70[472:TTHMK]2.0.CO;2. S2CID 198969439.
- ^ a b Cox, Cymon J.; Foster, Peter G.; Hirt, Robert P.; Harris, Simon R.; Embley, T. Martin (2008). "The archaebacterial origin of eukaryotes". Proceedings of the National Academy of Sciences of the United States of America. 105 (51): 20356–20361. Bibcode:2008PNAS..10520356C. doi:10.1073/pnas.0810647105. PMC 2629343. PMID 19073919.
- ^ MacLeod, Fraser; Kindler, Gareth S.; Wong, Hon Lun; Chen, Ray; Burns, Brendan P. (2019). "Asgard archaea: Diversity, function, and evolutionary implications in a range of microbiomes". AIMS Microbiology. 5 (1): 48–61. doi:10.3934/microbiol.2019.1.48. PMC 6646929. PMID 31384702.
- ^ Jüttner, Michael; Ferreira-Cerca, Sébastien (2022). "Looking through the Lens of the Ribosome Biogenesis Evolutionary History: Possible Implications for Archaeal Phylogeny and Eukaryogenesis". Molecular Biology and Evolution. 39 (4): msac054. doi:10.1093/molbev/msac054. PMC 8997704. PMID 35275997.
- ^ Eme, Laura; Spang, Anja; Lombard, Jonathan; Stairs, Courtney W.; Ettema, Thijs J. G. (2017). "Archaea and the origin of eukaryotes". Nature Reviews Microbiology. 15 (12): 711–723. doi:10.1038/nrmicro.2017.133. PMID 29123225. S2CID 8666687.
- ^ Vishwanath, Prashanth; Favaretto, Paola; Hartman, Hyman; Mohr, Scott C.; Smith, Temple F. (2004). "Ribosomal protein-sequence block structure suggests complex prokaryotic evolution with implications for the origin of eukaryotes". Molecular Phylogenetics and Evolution. 33 (3): 615–625. doi:10.1016/j.ympev.2004.07.003. PMID 15522791.
- ^ Rivera, Maria C.; Lake, James A. (2004). "The ring of life provides evidence for a genome fusion origin of eukaryotes". Nature. 431 (7005): 152–155. Bibcode:2004Natur.431..152R. doi:10.1038/nature02848. PMID 15356622. S2CID 4349149.
- ^ Pisani, Davide; Cotton, James A.; McInerney, James O. (2007). "Supertrees disentangle the chimerical origin of eukaryotic genomes". Molecular Biology and Evolution. 24 (8): 1752–1760. doi:10.1093/molbev/msm095. PMID 17504772.
- ^ Yutin, Natalya; Makarova, Kira S.; Mekhedov, Sergey L.; Wolf, Yuri I.; Koonin, Eugene V. (2008). "The deep archaeal roots of eukaryotes". Molecular Biology and Evolution. 25 (8): 1619–1630. doi:10.1093/molbev/msn108. PMC 2464739. PMID 18463089.
- ^ Yutin, Natalya; Wolf, Maxim Y.; Wolf, Yuri I.; Koonin, Eugene V. (2009). "The origins of phagocytosis and eukaryogenesis". Biology Direct. 4: 9. doi:10.1186/1745-6150-4-9. PMC 2651865. PMID 19245710.
- ^ Yutin, Natalya; Koonin, Eugene V. (2012). "Archaeal origin of tubulin". Biology Direct. 7: 10. doi:10.1186/1745-6150-7-10. PMC 3349469. PMID 22458654.
- ^ Samson, Rachel Y.; Dobro, Megan J.; Jensen, Grant J.; Bell, Stephen D. (2017). "The Structure, Function and Roles of the Archaeal ESCRT Apparatus". Prokaryotic Cytoskeletons. Subcellular Biochemistry. Vol. 84. pp. 357–377. doi:10.1007/978-3-319-53047-5_12. ISBN 978-3-319-53045-1. PMID 28500532.
- ^ Pelve, Erik A.; Lindås, Ann-Christin; Martens-Habbena, Willm; de la Torre, José R.; Stahl, David A.; Bernander, Rolf (2011). "Cdv-based cell division and cell cycle organization in the thaumarchaeon Nitrosopumilus maritimus". Molecular Microbiology. 82 (3): 555–566. doi:10.1111/j.1365-2958.2011.07834.x. PMID 21923770. S2CID 1202516.
- ^ Dobro, Megan J.; Samson, Rachel Y.; Yu, Zhiheng; McCullough, John; Ding, H. Jane; Chong, Parkson Lee-Gau; Bell, Stephen D.; Jensen, Grant J. (2013). "Electron cryotomography of ESCRT assemblies and dividing Sulfolobus cells suggests that spiraling filaments are involved in membrane scission". Molecular Biology of the Cell. 24 (15): 2319–2327. doi:10.1091/mbc.E12-11-0785. PMC 3727925. PMID 23761076.
- ^ Grau-Bové, Xavier; Sebé-Pedrós, Arnau; Ruiz-Trillo, Iñaki (2015). "The eukaryotic ancestor had a complex ubiquitin signaling system of archaeal origin". Molecular Biology and Evolution. 32 (3): 726–739. doi:10.1093/molbev/msu334. PMC 4327156. PMID 25525215.
- ^ Makarova, Kira S.; Koonin, Eugene V. (2010). "Archaeal ubiquitin-like proteins: functional versatility and putative ancestral involvement in tRNA modification revealed by comparative genomic analysis". Archaea. 2010: 710303. doi:10.1155/2010/710303. PMC 2948915. PMID 20936112.
- ^ Makarova, Kira S.; Koonin, Eugene V.; Kelman, Zvi (2012). "The CMG (CDC45/RecJ, MCM, GINS) complex is a conserved component of the DNA replication system in all archaea and eukaryotes". Biology Direct. 7: 7. doi:10.1186/1745-6150-7-7. PMC 3307487. PMID 22329974.
- ^ Stairs, Courtney W.; Ettema, Thijs J.G. (2020). "The Archaeal Roots of the Eukaryotic Dynamic Actin Cytoskeleton". Current Biology. 30 (10): R521–R526. doi:10.1016/j.cub.2020.02.074. PMID 32428493. S2CID 218710903.
- ^ a b Fournier, Gregory P.; Poole, Anthony M. (2018). "A Briefly Argued Case That Asgard Archaea Are Part of the Eukaryote Tree". Frontiers in Microbiology. 9: 1896. doi:10.3389/fmicb.2018.01896. PMC 6104171. PMID 30158917.
- ^ Jørgensen, Steffen Leth; Hannisdal, Bjarte; Lanzen, Anders; Baumberger, Tamara; Flesland, Kristin; Fonseca, Rita; Øvreås, Lise; Steen, Ida H; Thorseth, Ingunn H; Pedersen, Rolf B; Schleper, Christa (September 5, 2012). "Correlating microbial community profiles with geochemical data in highly stratified sediments from the Arctic Mid-Ocean Ridge". PNAS. 109 (42): E2846–55. doi:10.1073/pnas.1207574109. PMC 3479504. PMID 23027979.
- ^ Jørgensen, Steffen Leth; Thorseth, Ingunn H; Pedersen, Rolf B; Baumberger, Tamara; Schleper, Christa (October 4, 2013). "Quantitative and phylogenetic study of the Deep Sea Archaeal Group in sediments of the Arctic mid-ocean spreading ridge". Frontiers in Microbiology. 4: 299. doi:10.3389/fmicb.2013.00299. PMC 3790079. PMID 24109477.
- ^ a b Spang, Anja; Saw, Jimmy H.; Jørgensen, Steffen L.; Zaremba-Niedzwiedzka, Katarzyna; Martijn, Joran; Lind, Anders E.; Eijk, Roel van; Schleper, Christa; Guy, Lionel (2015). "Complex archaea that bridge the gap between prokaryotes and eukaryotes". Nature. 521 (7551): 173–179. Bibcode:2015Natur.521..173S. doi:10.1038/nature14447. PMC 4444528. PMID 25945739.
- ^ Da Cunha, Violette; Gaia, Morgan; Gadelle, Daniele; Nasir, Arshan; Forterre, Patrick (2017). "Lokiarchaea are close relatives of Euryarchaeota, not bridging the gap between prokaryotes and eukaryotes". PLOS Genetics. 13 (6): e1006810. doi:10.1371/journal.pgen.1006810. PMC 5484517. PMID 28604769.
- ^ Da Cunha, Violette; Gaïa, Morgan; Forterre, Patrick (2022). "The expanding Asgard archaea and their elusive relationships with Eukarya". mLife. 1 (1): 3–12. doi:10.1002/mlf2.12012. S2CID 247689333.
- ^ Da Cunha, Violette; Gaia, Morgan; Nasir, Arshan; Forterre, Patrick (2018). "Asgard archaea do not close the debate about the universal tree of life topology". PLOS Genetics. 14 (3): e1007215. doi:10.1371/journal.pgen.1007215. ISSN 1553-7404. PMC 5875737. PMID 29596428.
- ^ Spang, Anja; Eme, Laura; Saw, Jimmy H.; Caceres, Eva F.; Zaremba-Niedzwiedzka, Katarzyna; Lombard, Jonathan; Guy, Lionel; Ettema, Thijs J. G. (2018). "Asgard archaea are the closest prokaryotic relatives of eukaryotes". PLOS Genetics. 14 (3): e1007080. doi:10.1371/journal.pgen.1007080. PMC 5875740. PMID 29596421.
- ^ Liu, Yang; Makarova, Kira S.; Huang, Wen-Cong; Wolf, Yuri I.; Nikolskaya, Anastasia N.; Zhang, Xinxu; Cai, Mingwei; Zhang, Cui-Jing; Xu, Wei; Luo, Zhuhua; Cheng, Lei (2021). "Expanded diversity of Asgard archaea and their relationships with eukaryotes". Nature. 593 (7860): 553–557. Bibcode:2021Natur.593..553L. doi:10.1038/s41586-021-03494-3. ISSN 0028-0836. PMID 33911286. S2CID 233447651.
- ^ Xie, Ruize; Wang, Yinzhao; Huang, Danyue; Hou, Jialin; Li, Liuyang; Hu, Haining; Zhao, Xiaoxiao; Wang, Fengping (2022). "Expanding Asgard members in the domain of Archaea sheds new light on the origin of eukaryotes". Science China Life Sciences. 65 (4): 818–829. doi:10.1007/s11427-021-1969-6. PMID 34378142. S2CID 236977853.
- ^ López-García, Purificación; Moreira, David (2020). "Cultured Asgard Archaea Shed Light on Eukaryogenesis". Cell. 181 (2): 232–235. doi:10.1016/j.cell.2020.03.058. PMID 32302567. S2CID 215798394.
- ^ Akıl, Caner; Robinson, Robert C. (2018). "Genomes of Asgard archaea encode profilins that regulate actin". Nature. 562 (7727): 439–443. Bibcode:2018Natur.562..439A. doi:10.1038/s41586-018-0548-6. PMID 30283132. S2CID 52917038.
- ^ Hennell James, Rory; Caceres, Eva F.; Escasinas, Alex; Alhasan, Haya; Howard, Julie A.; Deery, Michael J.; Ettema, Thijs J. G.; Robinson, Nicholas P. (2017). "Functional reconstruction of a eukaryotic-like E1/E2/(RING) E3 ubiquitylation cascade from an uncultured archaeon". Nature Communications. 8 (1): 1120. Bibcode:2017NatCo...8.1120H. doi:10.1038/s41467-017-01162-7. ISSN 2041-1723. PMC 5654768. PMID 29066714.
- ^ Klinger, Christen M.; Spang, Anja; Dacks, Joel B.; Ettema, Thijs J. G. (2016). "Tracing the Archaeal Origins of Eukaryotic Membrane-Trafficking System Building Blocks". Molecular Biology and Evolution. 33 (6): 1528–1541. doi:10.1093/molbev/msw034. PMID 26893300.
- ^ Akıl, Caner; Tran, Linh T.; Orhant-Prioux, Magali; Baskaran, Yohendran; Manser, Edward; Blanchoin, Laurent; Robinson, Robert C. (2020). "Insights into the evolution of regulated actin dynamics via characterization of primitive gelsolin/cofilin proteins from Asgard archaea". Proceedings of the National Academy of Sciences. 117 (33): 19904–19913. Bibcode:2020PNAS..11719904A. doi:10.1073/pnas.2009167117. PMC 7444086. PMID 32747565.
- ^ Liu, Yang; Makarova, Kira S.; Huang, Wen-Cong; Wolf, Yuri I.; Nikolskaya, Anastasia N.; Zhang, Xinxu; Cai, Mingwei; Zhang, Cui-Jing; Xu, Wei; Luo, Zhuhua; Cheng, Lei (2021). "Expanded diversity of Asgard archaea and their relationships with eukaryotes". Nature. 593 (7860): 553–557. Bibcode:2021Natur.593..553L. doi:10.1038/s41586-021-03494-3. PMID 33911286. S2CID 233447651.
- ^ Hugenholtz, Philip (2002). "Exploring prokaryotic diversity in the genomic era". Genome Biology. 3 (2): reviews0003.1. doi:10.1186/gb-2002-3-2-reviews0003. PMC 139013. PMID 11864374.
- ^ Oren, Aharon (2004). Godfray, H. C. J.; Knapp, S. (eds.). "Prokaryote diversity and taxonomy: current status and future challenges". Philosophical Transactions of the Royal Society of London. Series B: Biological Sciences. 359 (1444): 623–638. doi:10.1098/rstb.2003.1458. PMC 1693353. PMID 15253349.
- ^ Moore, Kelsey R.; Magnabosco, Cara; Momper, Lily; Gold, David A.; Bosak, Tanja; Fournier, Gregory P. (2019). "An Expanded Ribosomal Phylogeny of Cyanobacteria Supports a Deep Placement of Plastids". Frontiers in Microbiology. 10: 1612. doi:10.3389/fmicb.2019.01612. ISSN 1664-302X. PMC 6640209. PMID 31354692.
- ^ Seitz, Patrick; Blokesch, Melanie (2013). "Cues and regulatory pathways involved in natural competence and transformation in pathogenic and environmental Gram-negative bacteria". FEMS Microbiology Reviews. 37 (3): 336–363. doi:10.1111/j.1574-6976.2012.00353.x. PMID 22928673. S2CID 30861416.
- ^ Bibb, Mervyn J. (2013). "Understanding and manipulating antibiotic production in actinomycetes". Biochemical Society Transactions. 41 (6): 1355–1364. doi:10.1042/BST20130214. PMID 24256223.
- ^ Woodruff, H. Boyd (2014). "Selman A. Waksman, winner of the 1952 Nobel Prize for physiology or medicine". Applied and Environmental Microbiology. 80 (1): 2–8. Bibcode:2014ApEnM..80....2W. doi:10.1128/AEM.01143-13. PMC 3911012. PMID 24162573.
- ^ Schleifer, Karl Heinz (2009). "Classification of Bacteria and Archaea: Past, present and future". Systematic and Applied Microbiology. 32 (8): 533–542. doi:10.1016/j.syapm.2009.09.002. PMID 19819658.
- ^ Adl, Sina M.; Simpson, Alastair G. B.; Farmer, Mark A.; Andersen, Robert A.; Anderson, O. Roger; Barta, John R.; Bowser, Samuel S.; Brugerolle, Guy; Fensome, Robert A.; Fredericq, Suzanne; James, Timothy Y. (2005). "The new higher level classification of eukaryotes with emphasis on the taxonomy of protists". The Journal of Eukaryotic Microbiology. 52 (5): 399–451. doi:10.1111/j.1550-7408.2005.00053.x. PMID 16248873. S2CID 8060916.