Dip pen nanolithography (DPN) is a scanning probe lithography technique where an atomic force microscope (AFM) tip is used to directly create patterns on a substrate.[1] It can be done on a range of substances with a variety of inks. A common example of this technique is exemplified by the use of alkane thiolates to imprint onto a gold surface.[2] This technique allows surface patterning on scales of under 100 nanometers. DPN is the nanotechnology analog of the dip pen (also called the quill pen), where the tip of an atomic force microscope cantilever acts as a "pen", which is coated with a chemical compound or mixture acting as an "ink", and put in contact with a substrate, the "paper".[3]
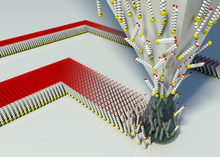
DPN enables direct deposition of nanoscale materials onto a substrate in a flexible manner. Recent advances have demonstrated massively parallel patterning using two-dimensional arrays of 55,000 tips.
Applications of this technology currently range through chemistry, materials science, and the life sciences, and include such work as ultra high density biological nanoarrays, and additive photomask repair.[4]
Development
editThe uncontrollable transfer of a molecular "ink" from a coated AFM tip to a substrate was first reported by Jaschke and Butt in 1995,[5] but they erroneously concluded that alkanethiols could not be transferred to gold substrates to form stable nanostructures. A research group at Northwestern University, US led by Chad Mirkin independently studied the process and determined that under the appropriate conditions, molecules could be transferred to a wide variety of surfaces to create stable chemically-adsorbed monolayers in a high resolution lithographic process they termed "DPN".[6] Mirkin and his coworkers hold the patents on this process,[7] and the patterning technique has expanded to include liquid "inks". It is important to note that "liquid inks" are governed by a very different deposition mechanism when compared to "molecular inks".
Deposition materials
editMolecular inks
editMolecular inks are typically composed of small molecules that are coated onto a DPN tip and are delivered to the surface through a water meniscus.[citation needed] In order to coat the tips, one can either vapor coat the tip or dip the tips into a dilute solution containing the molecular ink. If one dip-coats the tips, the solvent must be removed prior to deposition. The deposition rate of a molecular ink is dependent on the diffusion rate of the molecule, which is different for each molecule. The size of the feature is controlled by the tip/surface dwell-time (ranging from milliseconds to seconds) and the size of the water meniscus, which is determined by the humidity conditions (assuming the tip's radius of curvature is much smaller than the meniscus).
- Water meniscus mediated (exceptions do exist)
- Nanoscale feature resolution (50 nm to 2000 nm)
- No multiplexed depositions
- Each molecular ink is limited to its corresponding substrate
Examples
editLiquid inks
editLiquid inks can be any material that is liquid at deposition conditions. The liquid deposition properties are determined by the interactions between the liquid and the tip, the liquid and the surface, and the viscosity of the liquid itself. These interactions limit the minimum feature size of the liquid ink to about 1 micrometre, depending on the contact angle of the liquid. Higher viscosities offer greater control over feature size and are desirable. Unlike molecular inks, it is possible to perform multiplexed depositions using a carrier liquid. For example, using a viscous buffer, it is possible to directly deposit multiple proteins simultaneously.
- 1–10 micrometre feature resolution
- Multiplexed depositions
- Less restrictive ink/surface requirements
- Direct deposition of high viscosity materials
Examples
editApplications
editIn order to define a good DPN application, it is important to understand what DPN can do that other techniques cannot. Direct-write techniques, like contact printing, can pattern multiple biological materials but it cannot create features with subcellular resolution. Many high-resolution lithography methods can pattern at sub-micrometre resolution, but these require high-cost equipment that were not designed for biomolecule deposition and cell culture. Microcontact printing can print biomolecules at ambient conditions, but it cannot pattern multiple materials with nanoscale registry.
Industrial applications
editThe following are some examples of how DPN is being applied to potential products.
- Biosensor Functionalization – Directly place multiple capture domains on a single biosensor device
- Nanoscale Sensor Fabrication – Small, high-value sensors that can detect multiple targets[16]
- Nanoscale Protein Chips – High-density protein arrays with increased sensitivity
Emerging applications
editCell engineering
editDPN is emerging as a powerful research tool for manipulating cells at subcellular resolution[17][18]
- Stem cell differentiation
- Subcellular drug delivery
- Cell sorting
- Surface gradients
- Subcellular ECM protein patterns
- Cell adhesion
Rapid prototyping
edit- Plasmonics and Metamaterials
- Cell and tissue screening
Properties
editDirect write
editDPN is a direct write technique so it can be used for top-down and bottom-up lithography applications. In top-down work, the tips are used to deliver an etch resist to a surface, which is followed by a standard etching process.[19] In bottom-up applications, the material of interest is delivered directly to the surface via the tips.
Unique advantages
edit- Directed Placement – Directly print various materials onto existing nano and microstructures with nanoscale registry
- Direct Write – Maskless creation of arbitrary patterns with feature resolutions from as small as 50 nm and as large as 10 micrometres[20]
- Biocompatible – Subcellular to nanoscale resolution at ambient deposition conditions
- Scalable – Force independent, allowing for parallel depositions[21]
Thermal dip pen lithography
editA heated probe tip version of Dip Pen Lithography has also been demonstrated, thermal Dip Pen Lithography (tDPL), to deposit nanoparticles.[22] Semiconductor, magnetic, metallic, or optically active nanoparticles can be written to a substrate via this method. The particles are suspended in a Poly(methyl methacrylate) (PMMA) or equivalent polymer matrix, and heated by the probe tip until they begin to flow. The probe tip acts as a nano-pen, and can pattern nanoparticles into a programmed structure. Depending on the size of the nanoparticles, resolutions of 78–400 nm were attained. An O2 plasma etch can be used to remove the PMMA matrix, and in the case of Iron Oxide nanoparticles, further reduce the resolution of lines to 10 nm.[22] Advantages unique to tDPL are that it is a maskless additive process that can achieve very narrow resolutions, it can also easily write many types of nanoparticles without requiring special solution preparation techniques. However there are limitations to this method. The nanoparticles must be smaller than the radius of gyration of the polymer, in the case of PMMA this is about 6 nm. Additionally, as nanoparticles increase in size viscosity increases, slowing the process. For a pure polymer deposition speeds of 200 μm/s are achievable. Adding nanoparticles reduces speeds to 2 μm/s, but is still faster than regular Dip Pen Lithography.[22]
Beam pen lithography
editA two dimensional array of (PDMS) deformable transparent pyramid shaped tips are coated with an opaque layer of metal. The metal is then removed from the very tip of the pyramid, leaving an aperture for light to pass through. The array is then scanned across a surface and light is directed to the base of each pyramid via a micromirror array, which funnels the light toward the tip. Depending on the distance between the tips and the surface, light interacts with the surface in a near-field or far-field fashion, allowing sub-diffraction scale features (100 nm features with 400 nm light) or larger features to be fabricated. [23]
Common misconceptions
editDirect comparisons to other techniques
editThis article is written like a personal reflection, personal essay, or argumentative essay that states a Wikipedia editor's personal feelings or presents an original argument about a topic. (December 2023) |
The criticism most often directed at DPN is the patterning speed. The reason for this has more to do with how it is compared to other techniques rather than any inherent weaknesses. For example, the soft lithography method, microcontact printing (μCP), is the current standard for low cost, bench-top micro and nanoscale patterning, so it is easy to understand why DPN is compared directly to microcontact printing. The problem is that the comparisons are usually based upon applications that are strongly suited to μCP, instead of comparing them to some neutral application. μCP has the ability to pattern one material over a large area in a single stamping step, just as photolithography can pattern over a large area in a single exposure. Of course DPN is slow when it is compared to the strength of another technique. DPN is a maskless direct write technique that can be used to create multiple patterns of varying size, shape, and feature resolution, all on a single substrate. No one would try to apply microcontact printing to such a project because then it would never be worth the time and money required to fabricate each master stamp for each new pattern. Even if they did, microcontact printing would not be capable of aligning multiple materials from multiple stamps with nanoscale registry.[24] The best way to understand this misconception is to think about the different ways to apply photolithography and e-beam lithography. No one would try to use e-beam to solve a photolithography problem and then claim e-beam to be "too slow". Directly compared to photolithography's large area patterning capabilities, e-beam lithography is slow and yet, e-beam instruments can be found in every lab and nanofab in the world. The reason for this is because e-beam has unique capabilities that cannot be matched by photolithography, just as DPN has unique capabilities that cannot be matched by microcontact printing.
Connection to atomic force microscopy
editDPN evolved directly from AFM so it is not a surprise that people often assume that any commercial AFM can perform DPN experiments. In fact, DPN does not require an AFM, and an AFM does not necessarily have real DPN capabilities. There is an excellent analogy with scanning electron microscopy (SEM) and electron beam (E-beam) lithography. E-beam evolved directly from SEM technology and both use a focused electron beam, but it is not possible to perform modern E-beam lithography experiments on a SEM that lacks the proper lithography hardware and software components.
It is also important to consider one of the unique characteristics of DPN, namely its force independence. With virtually all ink/substrate combinations, the same feature size will be patterned no matter how hard the tip is pressing down against the surface.[25] As long as robust SiN tips are used, there is no need for complicated feedback electronics, no need for lasers, no need for quad photo-diodes, and no need for an AFM.
See also
editReferences
edit- ^ Ginger, David S.; Zhang, Hua; Mirkin, Chad A. (2004). "The Evolution of Dip-Pen Nanolithography". Angewandte Chemie International Edition. 43 (1): 30–45. doi:10.1002/anie.200300608. ISSN 1433-7851. PMID 14694469.
- ^ Piner, R. D. (1999). ""Dip-Pen" Nanolithography". Science. 283 (5402): 661–663. doi:10.1126/science.283.5402.661. ISSN 0036-8075. PMID 9924019. S2CID 27011581.
- ^ "DPN – Northwestern – Intro". Northwestern University. Archived from the original on 12 June 2013. Retrieved 7 May 2013.
- ^ Elhadj, Selim; Chernov, Alexander A; De Yoreo, James J (13 February 2008). "Solvent-mediated repair and patterning of surfaces by AFM". Nanotechnology. 19 (10). IOP Publishing: 105304. Bibcode:2008Nanot..19j5304E. doi:10.1088/0957-4484/19/10/105304. ISSN 0957-4484. PMID 21817697. S2CID 22708216.
- ^ Jaschke, M.; Butt, H.-J. (1995). "Deposition of Organic Material by the Tip of a Scanning Force Microscope". Langmuir. 11 (4): 1061–1064. doi:10.1021/la00004a004.
- ^ Piner, R. D.; Zhu, J.; Xu, F.; Hong, S.; Mirkin, C. A. (1999). "Dip Pen Nanolithography". Science. 283 (5402): 661–663. doi:10.1126/science.283.5402.661. PMID 9924019. S2CID 27011581.
- ^ "Dip-Pen Nanolithography". Archived from the original on 12 June 2013. Retrieved 7 May 2013.
- ^ Lee, K.-B. (7 February 2002). "Protein Nanoarrays Generated By Dip-Pen Nanolithography". Science. 295 (5560). American Association for the Advancement of Science (AAAS): 1702–1705. Bibcode:2002Sci...295.1702L. doi:10.1126/science.1067172. ISSN 0036-8075. PMID 11834780. S2CID 1050903.
- ^ Lee, S. W.; Oh, B.-K.; Sanedrin, R. G.; Salaita, K.; Fujigaya, T.; Mirkin, C. A. (2 May 2006). "Biologically Active Protein Nanoarrays Generated Using Parallel Dip-Pen Nanolithography". Advanced Materials. 18 (9). Wiley: 1133–1136. Bibcode:2006AdM....18.1133L. doi:10.1002/adma.200600070. ISSN 0935-9648. S2CID 136684662.
- ^ Sistiabudi, Rizaldi; Ivanisevic, Albena (2 October 2008). "Dip-Pen Nanolithography of Bioactive Peptides on Collagen-Terminated Retinal Membrane". Advanced Materials. 20 (19). Wiley: 3678–3681. Bibcode:2008AdM....20.3678S. doi:10.1002/adma.200800950. ISSN 0935-9648. S2CID 135957280.
- ^ Demers, L. M. (7 June 2002). "Direct Patterning of Modified Oligonucleotides on Metals and Insulators by Dip-Pen Nanolithography". Science. 296 (5574). American Association for the Advancement of Science (AAAS): 1836–1838. Bibcode:2002Sci...296.1836D. doi:10.1126/science.1071480. ISSN 0036-8075. PMID 12052950. S2CID 25516647.
- ^ Fu; Liu; Zhang; Dravid (2003). "Nanopatterning of "Hard" Magnetic Nanostructures via DPN and a Sol-based Ink". Nano Letters. 3 (6): 757–760. Bibcode:2003NanoL...3..757F. doi:10.1021/nl034172g. S2CID 44215721.
- ^ Su; Aslam; Fu; Wu; Dravid (2004). "Dip-pen nanopatterning of photosensitive conducting polymer using a monomer ink". Appl. Phys. Lett. 84 (21): 4200. Bibcode:2004ApPhL..84.4200S. doi:10.1063/1.1737469. S2CID 7050999.
- ^ Lenhert, Steven; Sun, Peng; Wang, Yuhuang; Fuchs, Harald; Mirkin, Chad A (2007). "Massively parallel dip-pen nanolithography of heterogeneous supported phospholipid multilayer patterns". Small. 3 (1): 71–75. doi:10.1002/smll.200600431. ISSN 1613-6810. PMID 17294472. S2CID 32133498.
- ^ Sekula, Sylwia; Fuchs, Jeanette; Weg-Remers, Susanne; Nagel, Peter; Schuppler, Stefan; et al. (2008). "Multiplexed Lipid Dip-Pen Nanolithography on Subcellular Scales for the Templating of Functional Proteins and Cell Culture". Small. 4 (10). Wiley: 1785–1793. doi:10.1002/smll.200800949. ISSN 1613-6810. PMID 18814174. S2CID 13843962.
- ^ Tang; Shi (2008). "Preparation of gas sensors via DPN". Sensors and Actuators B. 131 (2): 379–383. doi:10.1016/j.snb.2007.11.043.
- ^ Pulsipher, Abigail; Yousaf, Muhammad N. (2 March 2010). "Surface Chemistry and Cell Biological Tools for the Analysis of Cell Adhesion and Migration". ChemBioChem. 11 (6). Wiley: 745–753. doi:10.1002/cbic.200900787. ISSN 1439-4227. PMID 20198673. S2CID 8243543.
- ^ Yousaf, Muhammad N (2009). "Model substrates for studies of cell mobility". Current Opinion in Chemical Biology. 13 (5–6). Elsevier BV: 697–704. doi:10.1016/j.cbpa.2009.10.001. ISSN 1367-5931. PMID 19864174.
- ^ Zhang, Hua; Amro, Nabil A.; Disawal, Sandeep; Elghanian, Robert; Shile, Roger; Fragala, Joseph (2 January 2007). "High-Throughput Dip-Pen-Nanolithography-Based Fabrication of Si Nanostructures". Small. 3 (1). Wiley: 81–85. doi:10.1002/smll.200600393. ISSN 1613-6810. PMID 17294474.
- ^ Maskless lithography
- ^ Nature Chemistry Vol 1, August 2009
- ^ a b c Woo, Dai, King & Sheehan "Maskless Nanoscale Writing of Nanoparticle-Polymer Composites and Nanoparticle Assemblies using Thermal Nanoprobes" NanoLetters (2009)
- ^ Huo, Fengwei; Zheng, Gengfeng; Liao, Xing; Giam, Louise R.; Chai, Jinan; Chen, Xiaodong; Shim, Wooyoung; Mirkin, Chad A. (2010). "Beam pen lithography". Nature Nanotechnology. 5 (9): 637–640. Bibcode:2010NatNa...5..637H. doi:10.1038/nnano.2010.161. PMID 20676088.
- ^ Mei, Ying; Cannizzaro, Christopher; Park, Hyoungshin; Xu, Qiaobing; Bogatyrev, Said R.; Yi, Kevin; Goldman, Nathan; Langer, Robert; Anderson, Daniel G. (2008). "Cell-Compatible, Multicomponent Protein Arrays with Subcellular Feature Resolution". Small. 4 (10). Wiley: 1600–1604. doi:10.1002/smll.200800363. ISSN 1613-6810. PMC 2679812. PMID 18844310.
- ^ Exceptions exist when printing to soft materials – Maedler, C.; Chada, S.; Cui, X.; Taylor, M.; Yan, M.; La Rosa, A. (2008). "Creation of nanopatterns by local protonation of P4VP via dip pen nanolithography". Journal of Applied Physics. 104 (1): 014311–014311–4. Bibcode:2008JAP...104a4311M. doi:10.1063/1.2953090. ISSN 0021-8979. S2CID 120578436.